6UCA
 
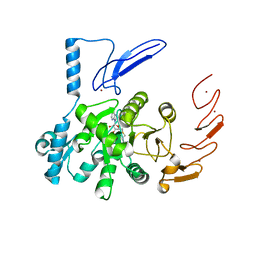 | | Crystal structure of human ZCCHC4 in complex with SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, rRNA N6-adenosine-methyltransferase ZCCHC4 | | Authors: | Lu, J.W, Ren, W.D, Huang, M.J, Gao, L, Li, D.X, Wang, G.G, Song, J. | | Deposit date: | 2019-09-15 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Structure and regulation of ZCCHC4 in m6A-methylation of 28S rRNA.
Nat Commun, 10, 2019
|
|
8UOT
 
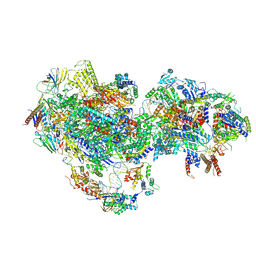 | | Composite map of PICdeltaTFIIK form1 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Yang, C, Murakami, K. | | Deposit date: | 2023-10-20 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Transcription start site scanning requires the fungi-specific hydrophobic loop of Tfb3.
Nucleic Acids Res., 2024
|
|
7BWE
 
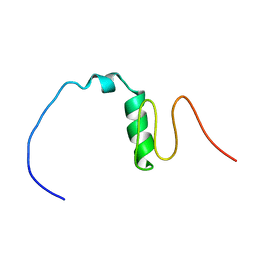 | | Consensus Chitin binding domain | | Descriptor: | Chitin binding beak protein 3 | | Authors: | Mohanram, H, Miserez, A. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a consensus chitin-binding domain revealed by solution NMR.
J.Struct.Biol., 213, 2021
|
|
6X39
 
 | |
8UMI
 
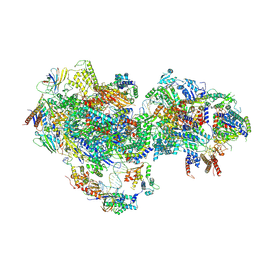 | | consensus map of PICdeltaTFIIK form1 | | Descriptor: | DNA (64-MER), DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Murakami, K. | | Deposit date: | 2023-10-17 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | consensus map of PICdeltaTFIIK form1
To be published
|
|
6UA4
 
 | | Human IMPDH2 treated with ATP, IMP, NAD+, and 2 mM GTP. Bent (3/4 compressed, 1/4 extended) segment reconstruction. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
6UE6
 
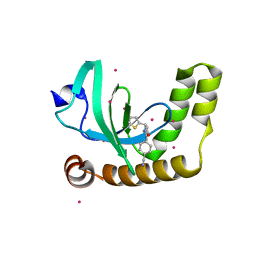 | | PWWP1 domain of NSD2 in complex with MR837 | | Descriptor: | 4-cyano-N-cyclopropyl-N-[(thiophen-2-yl)methyl]benzamide, Histone-lysine N-methyltransferase NSD2, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, De Freitas, R.F, Schapira, M, Brown, P.J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-09-20 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of Small-Molecule Antagonists of the PWWP Domain of NSD2.
J.Med.Chem., 64, 2021
|
|
8UMH
 
 | | Consensus map of PICdeltaTFIIK form2 | | Descriptor: | DNA (63-MER), DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Murakami, K. | | Deposit date: | 2023-10-17 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | consensus map of PICdeltaTFIIK form2
To be published
|
|
6WC6
 
 | |
6UG0
 
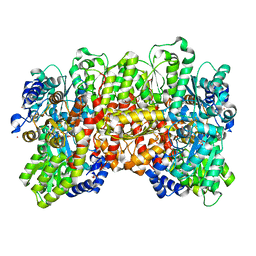 | | N2-bound Nitrogenase MoFe-protein from Azotobacter vinelandii | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Kang, W, Hu, Y, Ribbe, M.W. | | Deposit date: | 2019-09-25 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural evidence for a dynamic metallocofactor during N2reduction by Mo-nitrogenase.
Science, 368, 2020
|
|
5JYG
 
 | |
5K32
 
 | | PDE4D crystal structure in complex with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-[(3-methoxyphenyl)amino]-2-phenyl-7,8-dihydro-1,6-naphthyridin-5(6H)-one, MAGNESIUM ION, ... | | Authors: | Segarra, V, Hernandez, B, Roberts, R, Gracia, J, Soler, M, Bonin, I, Aymami, J. | | Deposit date: | 2016-05-19 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | 4-Amino-7,8-dihydro-1,6-naphthyridin-5(6 H)-ones as Inhaled Phosphodiesterase Type 4 (PDE4) Inhibitors: Structural Biology and Structure-Activity Relationships.
J. Med. Chem., 61, 2018
|
|
5JUT
 
 | | Saccharomyces cerevisiae 80S ribosome bound with elongation factor eEF2-GDP-sordarin and Taura Syndrome Virus IRES, Structure IV (almost non-rotated 40S subunit) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Abeyrathne, P, Koh, C.S, Grant, T, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-05-10 | | Release date: | 2016-10-05 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Ensemble cryo-EM uncovers inchworm-like translocation of a viral IRES through the ribosome.
Elife, 5, 2016
|
|
8W4P
 
 | | Structure of PSII-FCPII-I/J/K complex in the PSII-FCPII supercomplex from Cyclotella meneghiniana | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, CHLOROPHYLL A, ... | | Authors: | Shen, L.L, Li, Z.H, Shen, J.R, Wang, W.D. | | Deposit date: | 2023-08-24 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural insights into photosystem II supercomplex and trimeric FCP antennae of a centric diatom Cyclotella meneghiniana.
Nat Commun, 14, 2023
|
|
6TWQ
 
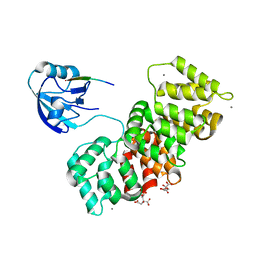 | | MAGI1_2 complexed with a 16E6 peptide | | Descriptor: | CALCIUM ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Gogl, G, Cousido-Siah, A, Trave, G. | | Deposit date: | 2020-01-13 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Dual Specificity PDZ- and 14-3-3-Binding Motifs: A Structural and Interactomics Study.
Structure, 28, 2020
|
|
8W4O
 
 | | Structure of PSII-FCPII-G/H complex in the PSII-FCPII supercomplex from Cyclotella meneghiniana | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15-octaen-17-ynyl]cyclohex-3-en-1-ol, (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, ... | | Authors: | Shen, L.L, Li, Z.H, Shen, J.R, Wang, W.D. | | Deposit date: | 2023-08-24 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural insights into photosystem II supercomplex and trimeric FCP antennae of a centric diatom Cyclotella meneghiniana.
Nat Commun, 14, 2023
|
|
7C89
 
 | |
7YN0
 
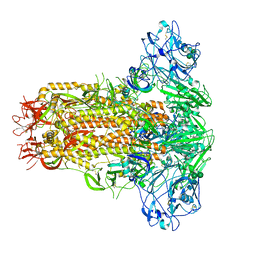 | | Cryo-EM structure of MERS-CoV spike protein, all RBD-down conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Chang, N.E, Weng, Z.W, Yang, T.J, Draczkowski, P. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Rapid simulation of glycoprotein structures by grafting and steric exclusion of glycan conformer libraries.
Cell, 187, 2024
|
|
6U8R
 
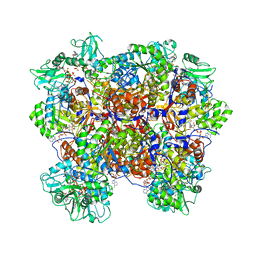 | | Human IMPDH2 treated with ATP, IMP, and NAD+. Bent (1/4 compressed, 3/4 extended) segment reconstruction. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 2, ... | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-05 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
7CW3
 
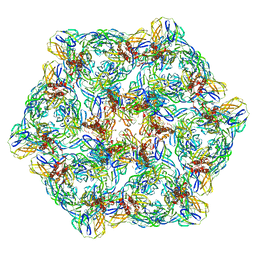 | | Cryo-EM structure of Chikungunya virus in complex with mAb CHK-263 IgG (subregion around icosahedral 2-fold vertex) | | Descriptor: | Capsid protein, E1 glycoprotein, E2 glycoprotein, ... | | Authors: | Zhou, Q.F, Fox, J.M, Earnest, J.T, Ng, T.S, Kim, A.S, Fibriansah, G, Kostyuchenko, V.A, Shu, B, Diamond, M.S, Lok, S.M. | | Deposit date: | 2020-08-27 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Structural basis of Chikungunya virus inhibition by monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8VL7
 
 | | Co-crystal structure of human TREX1 in complex with an inhibitor | | Descriptor: | (2P)-2-[3-bromo-2-(2-hydroxyethoxy)phenyl]-5-hydroxy-1-methyl-N-(1,2-oxazol-4-yl)-6-oxo-1,6-dihydropyrimidine-4-carboxamide, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Dehghani-Tafti, S, Dong, A, Li, Y, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L. | | Deposit date: | 2024-01-11 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Co-crystal structure of human TREX1 in complex with an inhibitor
To be published
|
|
6TVF
 
 | | Crystal structure of the haemagglutinin from a H10N7 seal influenza virus isolated in Germany in complex with human receptor analogue, 6'-SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Zhang, J, Xiong, X, Purkiss, A, Walker, P, Gamblin, S, Skehel, J.J. | | Deposit date: | 2020-01-09 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Hemagglutinin Traits Determine Transmission of Avian A/H10N7 Influenza Virus between Mammals.
Cell Host Microbe, 28, 2020
|
|
8VRM
 
 | | Crystal structure of the Pcryo_0619 N-acetyltransferase from Psychrobacter cryohalolentis K5 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, Acetyltransferase, ... | | Authors: | Dunsirn, M.M, Bockhaus, N.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2024-01-22 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Biochemical Investigation of the Enzymes Required for the Production of 2,3,4-triacetoamido-2,3,4-trideoxy-l-arabinose in Psychrobacter cryohalolentis K5
To Be Published
|
|
6WQR
 
 | |
6U0V
 
 | |
