3RBV
 
 | | Crystal Structure of KijD10, a 3-ketoreductase from Actinomadura kijaniata incomplex with NADP | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Holden, H.M, Kubiak, R.L. | | Deposit date: | 2011-03-30 | | Release date: | 2011-06-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Combined Structural and Functional Investigation of a C-3''-Ketoreductase Involved in the Biosynthesis of dTDP-l-Digitoxose.
Biochemistry, 50, 2011
|
|
4ZLI
 
 | | Cellobionic acid phosphorylase - 3-O-beta-D-glucopyranosyl-alpha-D-glucopyranuronic acid complex | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative b-glycan phosphorylase, ... | | Authors: | Nam, Y.W, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-05-01 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
5EQY
 
 | | Crystal structure of choline kinase alpha-1 bound by 5-[(4-methyl-1,4-diazepan-1-yl)methyl]-2-[4-[(4-methyl-1,4-diazepan-1-yl)methyl]phenyl]benzenecarbonitrile (compound 65) | | Descriptor: | 5-[(4-methyl-1,4-diazepan-1-yl)methyl]-2-[4-[(4-methyl-1,4-diazepan-1-yl)methyl]phenyl]benzenecarbonitrile, Choline kinase alpha | | Authors: | Zhou, T, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2015-11-13 | | Release date: | 2016-01-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel Small Molecule Inhibitors of Choline Kinase Identified by Fragment-Based Drug Discovery.
J.Med.Chem., 59, 2016
|
|
7DB4
 
 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in TDP012- and glutathione-bound form | | Descriptor: | 1-[(4-fluorophenyl)methyl]-2,2-bis(oxidanylidene)thieno[3,2-c][1,2]thiazin-4-one, GLUTATHIONE, Glutathione S-transferase E14 | | Authors: | Koiwai, K, Inaba, K, Yumoto, F, Senda, T, Niwa, R. | | Deposit date: | 2020-10-18 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Non-steroidal inhibitors of Drosophila melanogaster steroidogenic glutathione S -transferase Noppera-bo
J Pestic Sci, 46, 2021
|
|
3MHC
 
 | | Crystal structure of human cabonic anhydrase II in adduct with an adamantyl analogue of acetazolamide in a novel hydrophobic binding pocket | | Descriptor: | (3S,5S,7S)-N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)tricyclo[3.3.1.1~3,7~]decane-1-carboxamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Avvaru, B.S. | | Deposit date: | 2010-04-07 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Carbonic anhydrase inhibitors. The X-ray crystal structure of human isoform II in adduct with an adamantyl analogue of acetazolamide resides in a less utilized binding pocket than most hydrophobic inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6I53
 
 | | Cryo-EM structure of the human synaptic alpha1-beta3-gamma2 GABAA receptor in complex with Megabody38 in a lipid nanodisc | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-1, ... | | Authors: | Laverty, D, Desai, R, Uchanski, T, Masiulis, S, Wojciech, J.S, Malinauskas, T, Zivanov, J, Pardon, E, Steyaert, J, Miller, K.W, Aricescu, A.R. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the human alpha 1 beta 3 gamma 2 GABAAreceptor in a lipid bilayer.
Nature, 565, 2019
|
|
4Z3X
 
 | | Active site complex BamBC of Benzoyl Coenzyme A reductase in complex with 1-Monoenoyl-CoA | | Descriptor: | 1,5 Dienoyl-CoA, Benzoyl-CoA reductase, putative, ... | | Authors: | Weinert, T, Kung, J.W, Weidenweber, S, Huwiler, S.G, Boll, M, Ermler, U. | | Deposit date: | 2015-04-01 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of enzymatic benzene ring reduction.
Nat.Chem.Biol., 11, 2015
|
|
4PS7
 
 | |
3HM4
 
 | |
3ZX0
 
 | | NTPDase1 in complex with Heptamolybdate | | Descriptor: | ACETIC ACID, CHLORIDE ION, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE 1, ... | | Authors: | Zebisch, M, Schaefer, P, Straeter, N. | | Deposit date: | 2011-08-04 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic evidence for a domain motion in rat nucleoside triphosphate diphosphohydrolase (NTPDase) 1.
J. Mol. Biol., 415, 2012
|
|
1D1J
 
 | | CRYSTAL STRUCTURE OF HUMAN PROFILIN II | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, PROFILIN II, ... | | Authors: | Nodelman, I.M, Bowman, G.D, Lindberg, U, Schutt, C.E. | | Deposit date: | 1999-09-17 | | Release date: | 2000-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structure determination of human profilin II: A comparative structural analysis of human profilins.
J.Mol.Biol., 294, 1999
|
|
4JN9
 
 | | Crystal structure of the DepH | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DepH, ... | | Authors: | Li, J, Wang, C, Zhang, Z.M, Zhou, J.H, Cheng, E. | | Deposit date: | 2013-03-14 | | Release date: | 2014-04-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis of an NADP+-independent dithiol oxidase in FK228 biosynthesis.
Sci Rep, 4, 2014
|
|
3SOG
 
 | | Crystal structure of the BAR domain of human Amphiphysin, isoform 1 | | Descriptor: | 1,2-ETHANEDIOL, Amphiphysin, POTASSIUM ION | | Authors: | Allerston, C.K, Krojer, T, Chaikuad, A, Cooper, C.D.O, Berridge, G, Savitsky, P, Vollmar, M, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-30 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: |
|
|
7D9R
 
 | | Structure of huamn soluble guanylate cyclase in the riociguat and NO-bound state | | Descriptor: | Guanylate cyclase soluble subunit alpha-1, Guanylate cyclase soluble subunit beta-1, MAGNESIUM ION, ... | | Authors: | Chen, L, Liu, R, Kang, Y. | | Deposit date: | 2020-10-14 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Activation mechanism of human soluble guanylate cyclase by stimulators and activators.
Nat Commun, 12, 2021
|
|
7D9S
 
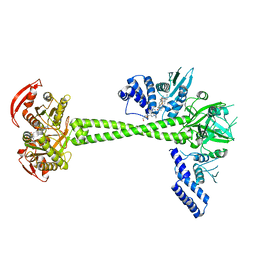 | | Structure of huamn soluble guanylate cyclase in the YC1 and NO-bound state | | Descriptor: | Guanylate cyclase soluble subunit alpha-1, Guanylate cyclase soluble subunit beta-1, MAGNESIUM ION, ... | | Authors: | Chen, L, Liu, R, Kang, Y. | | Deposit date: | 2020-10-14 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Activation mechanism of human soluble guanylate cyclase by stimulators and activators.
Nat Commun, 12, 2021
|
|
2V0K
 
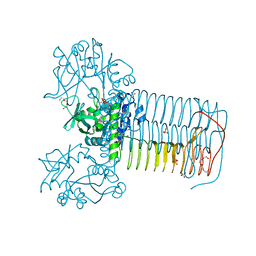 | | Characterization of Substrate Binding and Catalysis of the Potential Antibacterial Target N-acetylglucosamine-1-phosphate Uridyltransferase (GlmU) | | Descriptor: | BIFUNCTIONAL PROTEIN GLMU, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Mochalkin, I, Lightle, S, Ohren, J.F, Chirgadze, N.Y. | | Deposit date: | 2007-05-14 | | Release date: | 2008-01-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of Substrate Binding and Catalysis in the Potential Antibacterial Target N-Acetylglucosamine-1-Phosphate Uridyltransferase (Glmu).
Protein Sci., 16, 2007
|
|
4F32
 
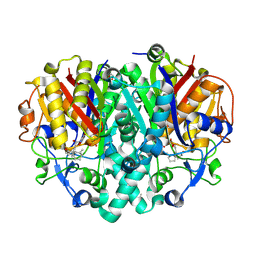 | | Crystal structure of 3-oxoacyl-[acyl-carrier-protein] synthase II from Burkholderia vietnamiensis in complex with platencin | | Descriptor: | 1,2-ETHANEDIOL, 2,4-dihydroxy-3-({3-[(2S,4aS,8S,8aR)-8-methyl-3-methylidene-7-oxo-1,3,4,7,8,8a-hexahydro-2H-2,4a-ethanonaphthalen-8-yl]propanoyl}amino)benzoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 2, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2012-05-08 | | Release date: | 2012-05-23 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Combining functional and structural genomics to sample the essential Burkholderia structome.
Plos One, 8, 2013
|
|
1CZF
 
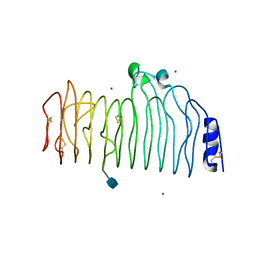 | | ENDO-POLYGALACTURONASE II FROM ASPERGILLUS NIGER | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, POLYGALACTURONASE II, ZINC ION | | Authors: | van Santen, Y, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 1999-09-02 | | Release date: | 1999-10-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | 1.68-A crystal structure of endopolygalacturonase II from Aspergillus niger and identification of active site residues by site-directed mutagenesis.
J.Biol.Chem., 274, 1999
|
|
4Z4Z
 
 | | Crystal structure of GII.10 P domain in complex with 30mM B antigen (trisaccharide) | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, alpha-L-fucopyranose-(1-2)-[alpha-D-galactopyranose-(1-3)]alpha-D-galactopyranose | | Authors: | Leuthold, M.M, Koromyslova, A.D, Hansman, G.S. | | Deposit date: | 2015-04-02 | | Release date: | 2015-05-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | The sweet quartet: Binding of fucose to the norovirus capsid.
Virology, 483, 2015
|
|
4J5B
 
 | | Human Cyclophilin D Complexed with an Inhibitor | | Descriptor: | 1-(4-aminobenzyl)-3-(2-{(2R)-2-[2-(methylsulfanyl)phenyl]pyrrolidin-1-yl}-2-oxoethyl)urea, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Gelin, M, Colliandre, L, Bessin, Y, Guichou, J.F. | | Deposit date: | 2013-02-08 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Fragment-based discovery of a new family of non-peptidic small-molecule cyclophilin inhibitors with potent antiviral activities.
Nat Commun, 7, 2016
|
|
1LPP
 
 | |
2VD6
 
 | | Human adenylosuccinate lyase in complex with its substrate N6-(1,2- Dicarboxyethyl)-AMP, and its products AMP and fumarate. | | Descriptor: | 2-[9-(3,4-DIHYDROXY-5-PHOSPHONOOXYMETHYL-TETRAHYDRO-FURAN-2-YL)-9H-PURIN-6-YLAMINO]-SUCCINIC ACID, ADENOSINE MONOPHOSPHATE, ADENYLOSUCCINATE LYASE, ... | | Authors: | Stenmark, P, Moche, M, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg-schiavone, L, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Nilsson, M, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Sundstrom, M, Thorsell, A.G, Tresaugues, L, van den Berg, S, Weigelt, J, Welin, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-09-30 | | Release date: | 2007-10-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human Adenylosuccinate Lyase in Complex with its Substrate N6-(1,2-Dicarboxyethyl)-AMP, and its Products AMP and Fumarate.
To be Published
|
|
4ZAK
 
 | | Crystal structure of the mCD1d/DB06-1/iNKTCR ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Birkholz, A.M. | | Deposit date: | 2015-04-13 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.824 Å) | | Cite: | A Novel Glycolipid Antigen for NKT Cells That Preferentially Induces IFN-gamma Production.
J Immunol., 195, 2015
|
|
1YIK
 
 | | Structure of Hen egg white lysozyme soaked with Cu-cyclam | | Descriptor: | 1,4,8,11-TETRAAZA-CYCLOTETRADECANE CU(II), ACETATE ION, CHLORIDE ION, ... | | Authors: | Hunter, T.M, McNae, I.W, Liang, X, Bella, J, Parsons, S, Walkinshaw, M.D, Sadler, P.J. | | Deposit date: | 2005-01-12 | | Release date: | 2005-02-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Protein recognition of macrocycles: binding of anti-HIV metallocyclams to lysozyme
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
4F66
 
 | | The crystal structure of 6-phospho-beta-glucosidase from Streptococcus mutans UA159 in complex with beta-D-glucose-6-phosphate. | | Descriptor: | 1,2-ETHANEDIOL, 6-O-phosphono-beta-D-glucopyranose, FORMIC ACID, ... | | Authors: | Tan, K, Michalska, K, Hatzos-Skintges, C, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-14 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.479 Å) | | Cite: | GH1-family 6-P-beta-glucosidases from human microbiome lactic acid bacteria.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
