5A89
 
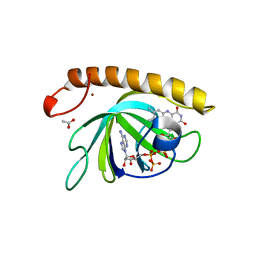 | | Crystal structure of the riboflavin kinase module of FAD synthetase from Corynebacterium ammoniagenes in complex with FMN and ADP(P 21 21 21) | | Descriptor: | ACETIC ACID, ADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Herguedas, B, Martinez-Julvez, M, Hermoso, J.A, Medina, M. | | Deposit date: | 2015-07-13 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Insights Into the Synthesis of Fmn in Prokaryotic Organisms.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3N8B
 
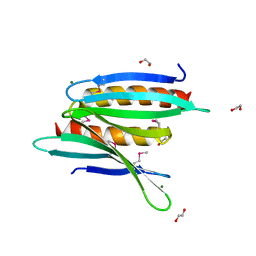 | | Crystal Structure of Borrelia burgdorferi Pur-alpha | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Uncharacterized protein | | Authors: | Graebsch, A, Roche, S, Kostrewa, D, Niessing, D. | | Deposit date: | 2010-05-28 | | Release date: | 2010-10-06 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Of bits and bugs--on the use of bioinformatics and a bacterial crystal structure to solve a eukaryotic repeat-protein structure.
Plos One, 5, 2010
|
|
1LJ8
 
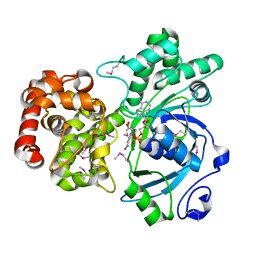 | | Crystal structure of mannitol dehydrogenase in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, mannitol dehydrogenase | | Authors: | Kavanagh, K.L, Klimacek, M, Nidetzky, B, Wilson, D.K. | | Deposit date: | 2002-04-19 | | Release date: | 2002-11-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Pseudomonas fluorescens mannitol 2-dehydrogenase binary and ternary
complexes. Specificity and catalytic mechanism
J.Biol.Chem., 277, 2002
|
|
3J15
 
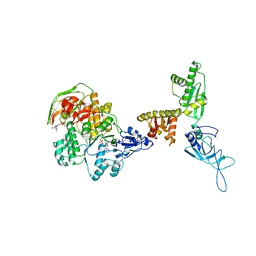 | | Model of ribosome-bound archaeal Pelota and ABCE1 | | Descriptor: | ABC transporter ATP-binding protein, ADENOSINE-5'-DIPHOSPHATE, IRON/SULFUR CLUSTER, ... | | Authors: | Becker, T, Franckenberg, S, Wickles, S, Shoemaker, C.J, Anger, A.M, Armache, J.-P, Sieber, H, Ungewickell, C, Berninghausen, O, Daberkow, I, Karcher, A, Thomm, M, Hopfner, K.-P, Green, R, Beckmann, R. | | Deposit date: | 2011-12-12 | | Release date: | 2012-02-22 | | Last modified: | 2018-08-22 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural basis of highly conserved ribosome recycling in eukaryotes and archaea.
Nature, 482, 2012
|
|
3O4B
 
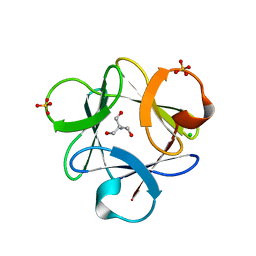 | |
3O4A
 
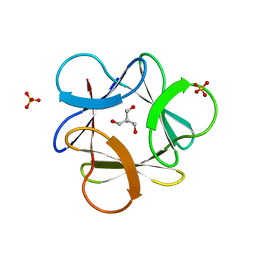 | |
8IT0
 
 | | Cryo-EM structure of Crt-SPARTA-gRNA-tDNA dimer (conformation-2) | | Descriptor: | DNA (45-mer), Piwi domain-containing protein, RNA (5'-R(P*UP*GP*AP*GP*GP*UP*AP*GP*UP*AP*GP*GP*UP*UP*GP*UP*AP*UP*AP*GP*U)-3'), ... | | Authors: | Gao, X, Shang, K, Zhu, K, Wang, L, Mu, Z, Fu, X, Yu, X, Qin, B, Zhu, H, Ding, W, Cui, S. | | Deposit date: | 2023-03-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Nature, 625, 2024
|
|
8ISZ
 
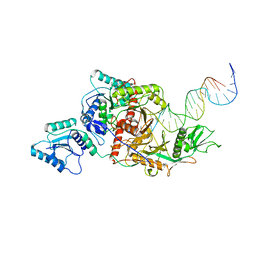 | | Cryo-EM structure of Crt-SPARTA-gRNA-tDNA monomer | | Descriptor: | DNA (45-mer), Piwi domain-containing protein, RNA (5'-R(P*UP*GP*AP*GP*GP*UP*AP*GP*UP*AP*GP*GP*UP*UP*GP*UP*AP*UP*AP*GP*U)-3'), ... | | Authors: | Gao, X, Shang, K, Zhu, K, Wang, L, Mu, Z, Fu, X, Yu, X, Qin, B, Zhu, H, Ding, W, Cui, S. | | Deposit date: | 2023-03-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Nature, 625, 2024
|
|
8ISY
 
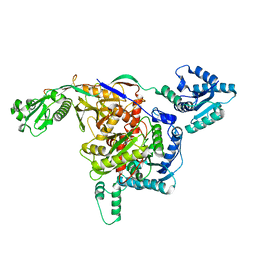 | | Cryo-EM structure of free-state Crt-SPARTA | | Descriptor: | Piwi domain-containing protein, TIR domain-containing protein | | Authors: | Gao, X, Shang, K, Zhu, K, Wang, L, Mu, Z, Fu, X, Yu, X, Qin, B, Zhu, H, Ding, W, Cui, S. | | Deposit date: | 2023-03-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Nature, 625, 2024
|
|
8IT1
 
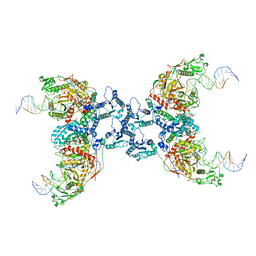 | | Cryo-EM structure of Crt-SPARTA-gRNA-tDNA tetramer (NADase active form) | | Descriptor: | DNA (45-mer), Piwi domain-containing protein, RNA (5'-R(P*UP*GP*AP*GP*GP*UP*AP*GP*UP*AP*GP*GP*UP*UP*GP*UP*AP*UP*AP*GP*U)-3'), ... | | Authors: | Gao, X, Shang, K, Zhu, K, Wang, L, Mu, Z, Fu, X, Yu, X, Qin, B, Zhu, H, Ding, W, Cui, S. | | Deposit date: | 2023-03-21 | | Release date: | 2023-11-08 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Nature, 625, 2024
|
|
8FFI
 
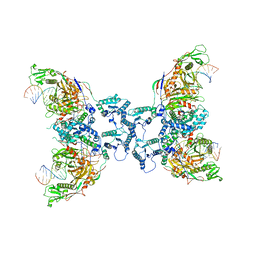 | | Structure of tetramerized MapSPARTA upon guide RNA-mediated target DNA binding | | Descriptor: | MAGNESIUM ION, TIR-APAZ, guide RNA, ... | | Authors: | Shen, Z.F, Yang, X.Y, Fu, T.M. | | Deposit date: | 2022-12-08 | | Release date: | 2023-08-23 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Oligomerization-mediated activation of a short prokaryotic Argonaute.
Nature, 621, 2023
|
|
8K9G
 
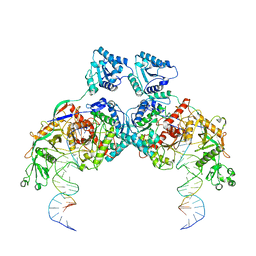 | | Cryo-EM structure of Crt-SPARTA-gRNA-tDNA dimer (conformation-1) | | Descriptor: | DNA (45-mer), Piwi domain-containing protein, RNA (5'-R(P*UP*GP*AP*GP*GP*UP*AP*GP*UP*AP*GP*GP*UP*UP*GP*UP*AP*UP*AP*GP*U)-3'), ... | | Authors: | Gao, X, Shang, K, Zhu, K, Wang, L, Mu, Z, Fu, X, Yu, X, Qin, B, Zhu, H, Ding, W, Cui, S. | | Deposit date: | 2023-08-01 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Nature, 625, 2024
|
|
7BEE
 
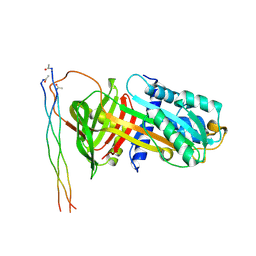 | |
7BDU
 
 | |
7U5G
 
 | | ACS122 Fab | | Descriptor: | ACS122 Fab Heavy chain, ACS122 Fab Light chain | | Authors: | Farokhi, E, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-03-02 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Complementary antibody lineages achieve neutralization breadth in an HIV-1 infected elite neutralizer.
Plos Pathog., 18, 2022
|
|
5EIR
 
 | | Co-crystal structure of eIF4E with nucleotide mimetic inhibitor. | | Descriptor: | Eukaryotic translation initiation factor 4 gamma 1, Eukaryotic translation initiation factor 4E, SULFATE ION, ... | | Authors: | Nowicki, M.W, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2015-10-30 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Design of nucleotide-mimetic and non-nucleotide inhibitors of the translation initiation factor eIF4E: Synthesis, structural and functional characterisation.
Eur.J.Med.Chem., 124, 2016
|
|
8I9O
 
 | | ecCTPS filament bound with CTP, NADH, DON | | Descriptor: | 5-OXO-L-NORLEUCINE, ADENINE, CTP synthase, ... | | Authors: | Guo, C.J, Liu, J.L. | | Deposit date: | 2023-02-07 | | Release date: | 2024-02-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Filamentation and inhibition of prokaryotic CTP synthase with ligands.
Mlife, 3, 2024
|
|
3UT2
 
 | | Crystal Structure of Fungal MagKatG2 | | Descriptor: | Catalase-peroxidase 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zamocky, M, Garcia-Fernandez, M.Q, Gasselhuber, B, Jakopitsch, C, Furtmuller, P.G, Loewen, P.C, Fita, I, Obinger, C, Carpena, X. | | Deposit date: | 2011-11-24 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | High Conformational Stability of Secreted Eukaryotic Catalase-peroxidases: ANSWERS FROM FIRST CRYSTAL STRUCTURE AND UNFOLDING STUDIES.
J.Biol.Chem., 287, 2012
|
|
3O4D
 
 | |
3O49
 
 | |
6CWX
 
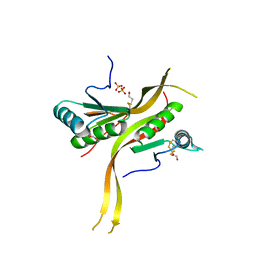 | | Crystal structure of human ribonuclease P/MRP proteins Rpp20/Rpp25 | | Descriptor: | FORMIC ACID, Ribonuclease P protein subunit p20, Ribonuclease P protein subunit p25, ... | | Authors: | Chan, C.W, Kiesel, B.R, Mondragon, A. | | Deposit date: | 2018-03-31 | | Release date: | 2018-04-18 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Human Rpp20/Rpp25 Reveals Quaternary Level Adaptation of the Alba Scaffold as Structural Basis for Single-stranded RNA Binding.
J. Mol. Biol., 430, 2018
|
|
4YKN
 
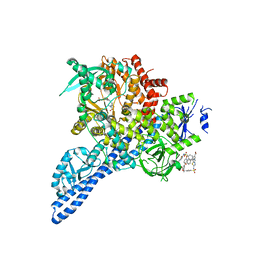 | | Pi3K alpha lipid kinase with Active Site Inhibitor | | Descriptor: | 3-(6-methoxypyridin-3-yl)-5-[({4-[(5-methyl-1,3,4-thiadiazol-2-yl)sulfamoyl]phenyl}amino)methyl]benzoic acid, Phosphatidylinositol 3-kinase regulatory subunit alpha,Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform fusion protein | | Authors: | Elkins, P.A. | | Deposit date: | 2015-03-04 | | Release date: | 2015-06-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of a Potent Class of PI3K alpha Inhibitors with Unique Binding Mode via Encoded Library Technology (ELT).
Acs Med.Chem.Lett., 6, 2015
|
|
7L6B
 
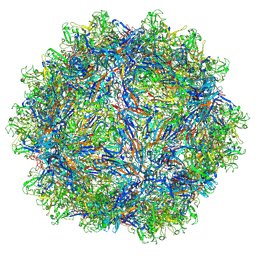 | | The empty AAV12 capsid | | Descriptor: | VP1 | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2020-12-23 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Completion of the AAV Structural Atlas: Serotype Capsid Structures Reveals Clade-Specific Features.
Viruses, 13, 2021
|
|
7L6A
 
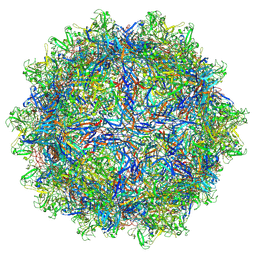 | | The genome-containing AAV12 capsid | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, VP1 | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2020-12-23 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Completion of the AAV Structural Atlas: Serotype Capsid Structures Reveals Clade-Specific Features.
Viruses, 13, 2021
|
|
7L6E
 
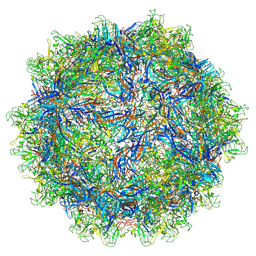 | | The genome-containing AAV11 capsid | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, Capsid protein | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2020-12-23 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Completion of the AAV Structural Atlas: Serotype Capsid Structures Reveals Clade-Specific Features.
Viruses, 13, 2021
|
|
