2WTP
 
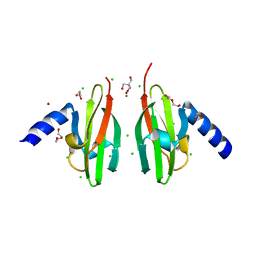 | | Crystal Structure of Cu-form Czce from C. metallidurans CH34 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Haertlein, I, Girard, E, Sarret, G, Hazemann, J, Gourhant, P, Kahn, R, Coves, J. | | Deposit date: | 2009-09-18 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Evidence for Conformational Changes Upon Copper Binding to Cupriavidus Metallidurans Czce.
Biochemistry, 49, 2010
|
|
4OWM
 
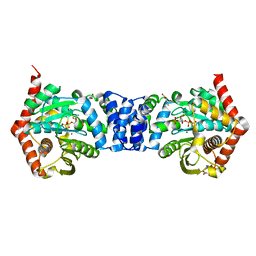 | | Anthranilate phosphoribosyl transferase from Mycobacterium tuberculosis in complex with 3-fluoroanthranilate, PRPP and Magnesium | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 2-azanyl-3-fluoranyl-benzoic acid, Anthranilate phosphoribosyltransferase, ... | | Authors: | Castell, A, Cookson, T.V.M, Short, F.L, Lott, J.S. | | Deposit date: | 2014-02-02 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Alternative substrates reveal catalytic cycle and key binding events in the reaction catalysed by anthranilate phosphoribosyltransferase from Mycobacterium tuberculosis.
Biochem.J., 461, 2014
|
|
4OF8
 
 | | Crystal Structure of Rst D1-D2 | | Descriptor: | GLYCEROL, Irregular chiasm C-roughest protein, SODIUM ION | | Authors: | Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-01-14 | | Release date: | 2014-02-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
4G5A
 
 | |
1P7T
 
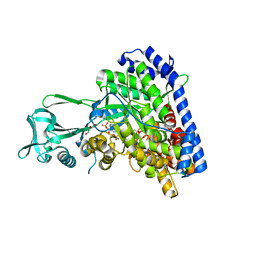 | | Structure of Escherichia coli malate synthase G:pyruvate:acetyl-Coenzyme A abortive ternary complex at 1.95 angstrom resolution | | Descriptor: | ACETYL COENZYME *A, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Anstrom, D.M, Kallio, K, Remington, S.J. | | Deposit date: | 2003-05-05 | | Release date: | 2003-09-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the Escherichia Coli Malate Synthase G:pyruvate:acetyl-coenzyme A Abortive Ternary Complex at 1.95 Angstrom Resolution
Protein Sci., 12, 2003
|
|
4JXJ
 
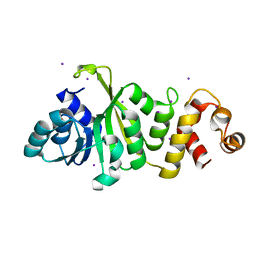 | |
4G8I
 
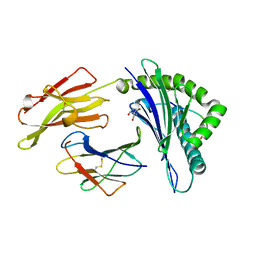 | | Crystal Structure of HLA B2705-KK10-L6M | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-2-microglobulin, Gag protein, ... | | Authors: | Gras, S, Wilmann, P.G, Rossjohn, J. | | Deposit date: | 2012-07-23 | | Release date: | 2013-03-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Molecular Basis for the Control of Preimmune Escape Variants by HIV-Specific CD8(+) T Cells.
Immunity, 38, 2013
|
|
2P8P
 
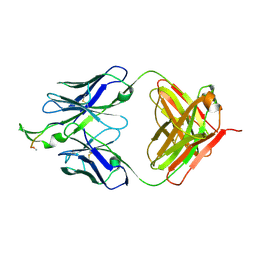 | | Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide LELDKWASLW[N-Ac] | | Descriptor: | gp41 peptide, nmAb 2F5, heavy chain, ... | | Authors: | Bryson, S, Julien, J.-P, Pai, E.F. | | Deposit date: | 2007-03-22 | | Release date: | 2007-05-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural details of HIV-1 recognition by the broadly neutralizing monoclonal antibody 2F5: epitope conformation, antigen-recognition loop mobility, and anion-binding site.
J.Mol.Biol., 384, 2008
|
|
3V1H
 
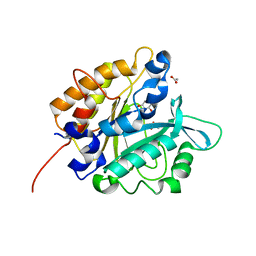 | | Structure of the H258Y mutant of Phosphatidylinositol-specific phospholipase C from Staphylococcus aureus | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1-phosphatidylinositol phosphodiesterase, ACETATE ION | | Authors: | Goldstein, R.I, Cheng, J, Stec, B, Roberts, M.F. | | Deposit date: | 2011-12-09 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the S. aureus PI-Specific Phospholipase C Reveals Modulation of Active Site Access by a Titratable PI-Cation Latched Loop
Biochemistry, 51, 2012
|
|
3GWI
 
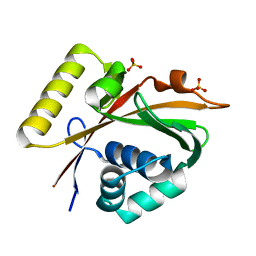 | | Crystal Structure of Mg-ATPase Nucleotide binding domain | | Descriptor: | Magnesium-transporting ATPase, P-type 1, SULFATE ION | | Authors: | Hakansson, K.O. | | Deposit date: | 2009-04-01 | | Release date: | 2009-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of Mg-ATPase nucleotide-binding domain at 1.6 A resolution reveals a unique ATP-binding motif
Acta Crystallogr.,Sect.D, 65, 2009
|
|
4TPY
 
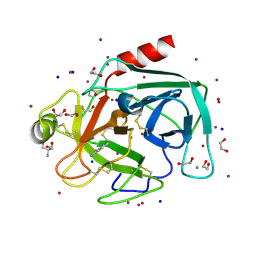 | | High throughput screening using acoustic droplet ejection to combine protein crystals and chemical libraries on crystallization plates at high density | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BENZAMIDINE, ... | | Authors: | Teplitsky, E, Joshi, K, Mullen, J.D, Soares, A.S. | | Deposit date: | 2014-06-10 | | Release date: | 2014-10-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High throughput screening using acoustic droplet ejection to combine protein crystals and chemical libraries on crystallization plates at high density.
J.Struct.Biol., 191, 2015
|
|
3FA6
 
 | |
2ZJF
 
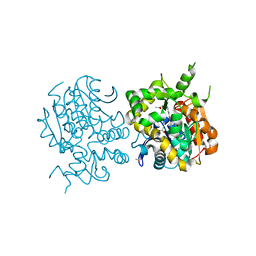 | |
2ZKG
 
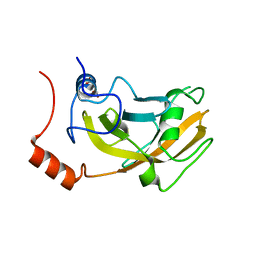 | | Crystal structure of unliganded SRA domain of mouse Np95 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase UHRF1 | | Authors: | Arita, K, Ariyoshi, M, Tochio, H, Nakamura, Y, Shirakawa, M. | | Deposit date: | 2008-03-19 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism
Nature, 455, 2008
|
|
2Z2R
 
 | | Nucleosome assembly proteins I (NAP-1, 74-365) | | Descriptor: | Nucleosome assembly protein | | Authors: | Park, Y.J, Luger, K. | | Deposit date: | 2007-05-25 | | Release date: | 2008-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A beta-hairpin comprising the nuclear localization sequence sustains the self-associated states of nucleosome assembly protein 1
J.Mol.Biol., 375, 2008
|
|
4BVG
 
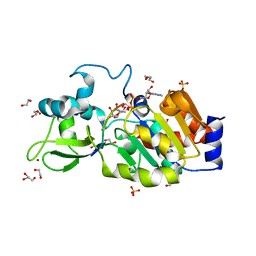 | | CRYSTAL STRUCTURE OF HUMAN SIRT3 IN COMPLEX WITH NATIVE ALKYLIMIDATE FORMED FROM ACETYL-LYSINE ACS2-PEPTIDE CRYSTALLIZED IN PRESENCE OF THE INHIBITOR EX-527 | | Descriptor: | (2R,3R,4S,5R)-5-({[(R)-{[(R)-{[(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]OXY}METHYL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL ACETATE, 1,2-ETHANEDIOL, ACETYL-COENZYME A SYNTHETASE 2-LIKE, ... | | Authors: | Nguyen, G.T.T, Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-06-25 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ex-527 inhibits Sirtuins by exploiting their unique NAD+-dependent deacetylation mechanism.
Proc. Natl. Acad. Sci. U.S.A., 110, 2013
|
|
2DZM
 
 | | Solution Structure of the Ubiquitin-like Domain in Human FAS-associated factor 1 (hFAF1) | | Descriptor: | FAS-associated factor 1 | | Authors: | Zhao, C, Sato, M, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-29 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Ubiquitin-like Domain in Human FAS-associated factor 1 (hFAF1)
To be Published
|
|
3AHJ
 
 | | H553A mutant of Phosphoketolase from Bifidobacterium Breve | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, THIAMINE DIPHOSPHATE, ... | | Authors: | Suzuki, R, Katayama, T, Kim, B.-J, Wakagi, T, Shoun, H, Ashida, H, Yamamoto, K, Fushinobu, S. | | Deposit date: | 2010-04-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of phosphoketolase: thiamine diphosphate-dependent dehydration mechanism
J.Biol.Chem., 285, 2010
|
|
1J5W
 
 | |
5P99
 
 | | rat catechol O-methyltransferase in complex with N-[2-[2-(3,4-dihydroxyphenyl)-2-oxoethyl]sulfanylethyl]-5-(4-fluorophenyl)-2,3-dihydroxybenzamide at 1.20A | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Ehler, A, Lerner, C, Rudolph, M.G. | | Deposit date: | 2016-08-30 | | Release date: | 2017-08-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structure of a COMT complex
To be published
|
|
4C69
 
 | | ATP binding to murine voltage-dependent anion channel 1 (mVDAC1). | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-5'-TRIPHOSPHATE, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Paz, A, Colletier, J.P, Abramson, J. | | Deposit date: | 2013-09-17 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.277 Å) | | Cite: | Structure-Guided Simulations Illuminate the Mechanism of ATP Transport Through Vdac1.
Nat.Struct.Mol.Biol., 21, 2014
|
|
3F8E
 
 | | Coumarins are a novel class of suicide carbonic anhydrase inhibitors | | Descriptor: | (2Z)-3-{2-hydroxy-5-[(1S)-1-hydroxy-3-methylbutyl]-4-methoxyphenyl}prop-2-enoic acid, BENZOIC ACID, Carbonic anhydrase 2, ... | | Authors: | Temperini, C, Maresca, A, Scozzafava, A, Supuran, C.T. | | Deposit date: | 2008-11-12 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Non-Zinc Mediated Inhibition of Carbonic Anhydrases: Coumarins Are a New Class of Suicide Inhibitors
J.Am.Chem.Soc., 131, 2009
|
|
3HO2
 
 | | Structure of E.coli FabF(C163A) in complex with Platencin | | Descriptor: | 2,4-dihydroxy-3-({3-[(2S,4aS,8S,8aR)-8-methyl-3-methylidene-7-oxo-1,3,4,7,8,8a-hexahydro-2H-2,4a-ethanonaphthalen-8-yl]propanoyl}amino)benzoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 2 | | Authors: | Soisson, S.M, Parthasarathy, G. | | Deposit date: | 2009-06-01 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Isolation, enzyme-bound structure and antibacterial activity of platencin A1 from Streptomyces platensis.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2JBP
 
 | | Protein kinase MK2 in complex with an inhibitor (crystal form-2, co- crystallization) | | Descriptor: | 2-(2-QUINOLIN-3-YLPYRIDIN-4-YL)-1,5,6,7-TETRAHYDRO-4H-PYRROLO[3,2-C]PYRIDIN-4-ONE, MAP KINASE-ACTIVATED PROTEIN KINASE 2 | | Authors: | Hillig, R.C, Eberspaecher, U, Monteclaro, F, Huber, M, Nguyen, D, Mengel, A, Muller-Tiemann, B, Egner, U. | | Deposit date: | 2006-12-09 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structural basis for a high affinity inhibitor bound to protein kinase MK2.
J. Mol. Biol., 369, 2007
|
|
4CXQ
 
 | | Mycobaterium tuberculosis transaminase BioA complexed with substrate KAPA | | Descriptor: | 1,2-ETHANEDIOL, 7-KETO-8-AMINOPELARGONIC ACID, ADENOSYLMETHIONINE-8-AMINO-7-OXONONANOATE AMINOTRANSFERASE, ... | | Authors: | Dai, R, Wilson, D.J, Geders, T.W, Aldrich, C.C, Finzel, B.C. | | Deposit date: | 2014-04-08 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition of Mycobacterium Tuberculosis Transaminase Bioa by Aryl Hydrazines and Hydrazides.
Chembiochem, 15, 2014
|
|
