8E3H
 
 | | Escherichia coli Rho-dependent transcription pre-termination complex containing 18 nt long RNA spacer, Mg-ADP-BeF3, and NusG; Rho hexamer part | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Molodtsov, V, Wang, C. | | Deposit date: | 2022-08-17 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Structural basis of Rho-dependent transcription termination.
Nature, 614, 2023
|
|
8E6W
 
 | | Escherichia coli Rho-dependent transcription pre-termination complex containing 18 nt long RNA spacer, lambda-tR1 rut RNA, Mg-ADP-BeF3, and NusG; Rho part | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H. | | Deposit date: | 2022-08-23 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Structural basis of Rho-dependent transcription termination.
Nature, 614, 2023
|
|
8E5P
 
 | | Escherichia coli Rho-dependent transcription pre-termination complex containing 24 nt long RNA spacer, Mg-ADP-BeF3, and NusG; Rho hexamer part | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Molodtsov, V, Wang, C. | | Deposit date: | 2022-08-22 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural basis of Rho-dependent transcription termination.
Nature, 614, 2023
|
|
8F2K
 
 | |
6LY8
 
 | | V/A-ATPase from Thermus thermophilus, the soluble domain, including V1, d, two EG stalks, and N-terminal domain of a-subunit. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, V-type ATP synthase alpha chain, V-type ATP synthase beta chain, ... | | Authors: | Kishikawa, J, Nakanishi, A, Furuta, A, Kato, T, Namba, K, Tamakoshi, M, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2020-02-13 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanical inhibition of isolated V o from V/A-ATPase for proton conductance.
Elife, 9, 2020
|
|
6N75
 
 | | Crystal Structure of ATPase delta1-79 Spa47 E287A | | Descriptor: | ATP synthase SpaL/MxiB | | Authors: | Morales, Y, Olsen, K.J, Johnson, S.J, Demler, H.J, Dickenson, N.E. | | Deposit date: | 2018-11-27 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.991 Å) | | Cite: | Interfacial amino acids support Spa47 oligomerization and shigella type three secretion system activation.
Proteins, 87, 2019
|
|
6N6L
 
 | | Crystal Structure of ATPase delta 1-79 Spa47 R189A R191A mutant | | Descriptor: | ATP synthase SpaL/MxiB, SULFATE ION | | Authors: | Morales, Y, Johnson, S.J, Demler, H.J, Dickenson, N.E. | | Deposit date: | 2018-11-26 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Interfacial amino acids support Spa47 oligomerization and shigella type three secretion system activation.
Proteins, 87, 2019
|
|
6N6Z
 
 | |
6N71
 
 | | Crystal Structure of ATPase delta1-79 Spa47 R191E | | Descriptor: | Probable ATP synthase SpaL/MxiB | | Authors: | Morales, Y, Olsen, K.J, Johnson, S.J, Demler, H.J, Dickenson, N.E. | | Deposit date: | 2018-11-27 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Interfacial amino acids support Spa47 oligomerization and shigella type three secretion system activation.
Proteins, 87, 2019
|
|
6N2Y
 
 | | Bacillus PS3 ATP synthase class 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Guo, H, Rubinstein, J.L. | | Deposit date: | 2018-11-14 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of a bacterial ATP synthase.
Elife, 8, 2019
|
|
6N74
 
 | | Crystal Structure of ATPase delta1-79 Spa47 R271E | | Descriptor: | ATP synthase SpaL/MxiB, SULFATE ION | | Authors: | Morales, Y, Johnson, S.J, Demler, H.J, Dickenson, N.E. | | Deposit date: | 2018-11-27 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Interfacial amino acids support Spa47 oligomerization and shigella type three secretion system activation.
Proteins, 87, 2019
|
|
6N70
 
 | | Crystal Structure of ATPase delta1-79 Spa47 R191A | | Descriptor: | ATP synthase SpaL/MxiB | | Authors: | Morales, Y, Olsen, K.J, Johnson, S.J, Demler, H.J, Dickenson, N.E. | | Deposit date: | 2018-11-27 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.738 Å) | | Cite: | Interfacial amino acids support Spa47 oligomerization and shigella type three secretion system activation.
Proteins, 87, 2019
|
|
6N6M
 
 | | Crystal Structure of ATPase delta1-79 Spa47 R189A | | Descriptor: | ATP synthase SpaL/MxiB, SULFATE ION | | Authors: | Morales, Y, Johnson, S.J, Demler, H.J, Dickenson, N.E. | | Deposit date: | 2018-11-26 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.788 Å) | | Cite: | Interfacial amino acids support Spa47 oligomerization and shigella type three secretion system activation.
Proteins, 87, 2019
|
|
6N2Z
 
 | | Bacillus PS3 ATP synthase class 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Guo, H, Rubinstein, J.L. | | Deposit date: | 2018-11-14 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of a bacterial ATP synthase.
Elife, 8, 2019
|
|
6N72
 
 | |
6N30
 
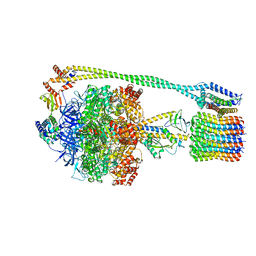 | | Bacillus PS3 ATP synthase class 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Guo, H, Rubinstein, J.L. | | Deposit date: | 2018-11-14 | | Release date: | 2019-02-20 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of a bacterial ATP synthase.
Elife, 8, 2019
|
|
6N76
 
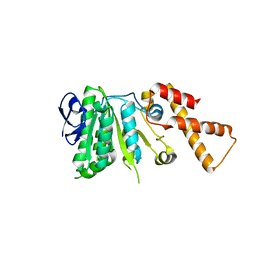 | |
6N73
 
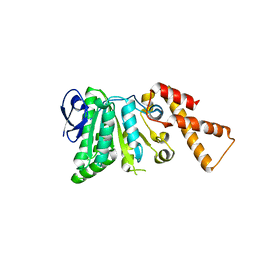 | |
6NJP
 
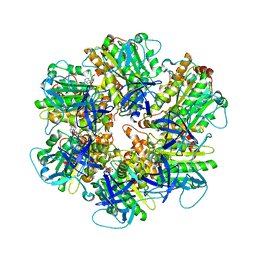 | | Structure of the assembled ATPase EscN in complex with its central stalk EscO from the enteropathogenic E. coli (EPEC) type III secretion system | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, EscO, ... | | Authors: | Majewski, D.D, Worrall, L.J, Hong, C, Atkinson, C.E, Vuckovic, M, Watanabe, N, Yu, Z, Strynadka, N.C.J. | | Deposit date: | 2019-01-03 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-EM structure of the homohexameric T3SS ATPase-central stalk complex reveals rotary ATPase-like asymmetry.
Nat Commun, 10, 2019
|
|
6NJO
 
 | | Structure of the assembled ATPase EscN from the enteropathogenic E. coli (EPEC) type III secretion system | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Majewski, D.D, Worrall, L.J, Hong, C, Atkinson, C.E, Vuckovic, M, Watanabe, N, Yu, Z, Strynadka, N.C.J. | | Deposit date: | 2019-01-03 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Cryo-EM structure of the homohexameric T3SS ATPase-central stalk complex reveals rotary ATPase-like asymmetry.
Nat Commun, 10, 2019
|
|
7KHR
 
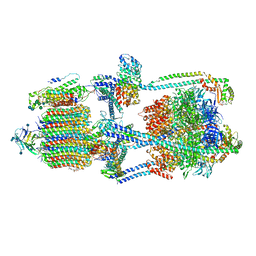 | | Cryo-EM structure of bafilomycin A1-bound intact V-ATPase from bovine brain | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (5R)-2,4-dideoxy-1-C-{(2S,3R,4S)-3-hydroxy-4-[(2R,3S,4E,6E,9R,10S,11R,12E,14Z)-10-hydroxy-3,15-dimethoxy-7,9,11,13-tetramethyl-16-oxo-1-oxacyclohexadeca-4,6,12,14-tetraen-2-yl]pentan-2-yl}-4-methyl-5-propan-2-yl-alpha-D-threo-pentopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, R, Li, X. | | Deposit date: | 2020-10-21 | | Release date: | 2021-03-17 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Molecular basis of V-ATPase inhibition by bafilomycin A1.
Nat Commun, 12, 2021
|
|
7L1S
 
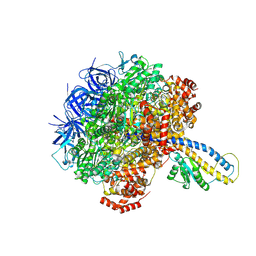 | | PS3 F1-ATPase Pi-bound Dwell | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ATP synthase subunit alpha, ... | | Authors: | Sobti, M, Ueno, H, Noji, H, Stewart, A.G. | | Deposit date: | 2020-12-15 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The six steps of the complete F 1 -ATPase rotary catalytic cycle.
Nat Commun, 12, 2021
|
|
7L1Q
 
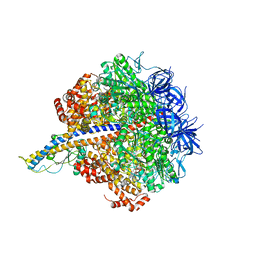 | | PS3 F1-ATPase Binding/TS Dwell | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Sobti, M, Ueno, H, Noji, H, Stewart, A.G. | | Deposit date: | 2020-12-15 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The six steps of the complete F 1 -ATPase rotary catalytic cycle.
Nat Commun, 12, 2021
|
|
7L1R
 
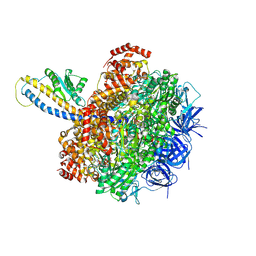 | | PS3 F1-ATPase Hydrolysis Dwell | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Sobti, M, Ueno, H, Noji, H, Stewart, A.G. | | Deposit date: | 2020-12-15 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The six steps of the complete F 1 -ATPase rotary catalytic cycle.
Nat Commun, 12, 2021
|
|
7NKJ
 
 | | Mycobacterium smegmatis ATP synthase F1 state 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Montgomery, M.G, Petri, J, Spikes, T.E, Walker, J.E. | | Deposit date: | 2021-02-18 | | Release date: | 2021-10-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | Structure of the ATP synthase from Mycobacterium smegmatis provides targets for treating tuberculosis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
