1L1R
 
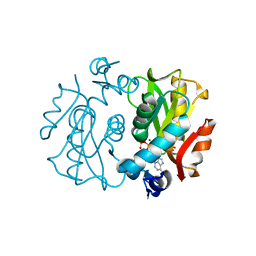 | | Crystal Structure of APRTase from Giardia lamblia Complexed with 9-deazaadenine, Mg2+ and PRPP | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 9-DEAZAADENINE, Adenine phosphoribosyltransferase, ... | | Authors: | Shi, W, Sarver, A.E, Wang, C.C, Tanaka, K.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2002-02-19 | | Release date: | 2002-11-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Closed Site Complexes of Adenine Phosphoribosyltransferase from Giardia lamblia
Reveal a Mechanism of Ribosyl Migration.
J.Biol.Chem., 277, 2002
|
|
4P3C
 
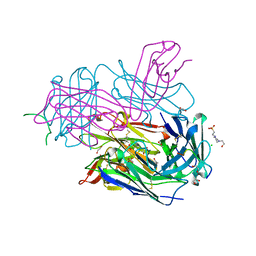 | | MT1-MMP:Fab complex (Form I) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Rozenberg, H, Udi, Y, Sagi, I. | | Deposit date: | 2014-03-06 | | Release date: | 2014-12-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.943 Å) | | Cite: | Inhibition mechanism of membrane metalloprotease by an exosite-swiveling conformational antibody.
Structure, 23, 2015
|
|
1L8C
 
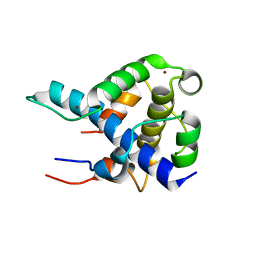 | | STRUCTURAL BASIS FOR HIF-1ALPHA/CBP RECOGNITION IN THE CELLULAR HYPOXIC RESPONSE | | Descriptor: | CREB-binding protein, Hypoxia-inducible factor 1 alpha, ZINC ION | | Authors: | Dames, S.A, Martinez-Yamout, M, De Guzman, R.N, Dyson, H.J, Wright, P.E. | | Deposit date: | 2002-03-19 | | Release date: | 2002-04-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for Hif-1 alpha /CBP recognition in the cellular hypoxic response.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
5DB0
 
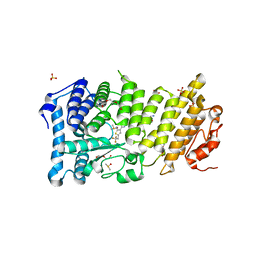 | | Menin in complex with MI-352 | | Descriptor: | 1-[(2R)-2,3-dihydroxypropyl]-5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1H-indole-2-carbonitrile, 1-[(2S)-2,3-dihydroxypropyl]-5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1 H-indole-2-carbonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Pollock, J, Dmitry, B, Cierpicki, T, Grembecka, J. | | Deposit date: | 2015-08-20 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Property Focused Structure-Based Optimization of Small Molecule Inhibitors of the Protein-Protein Interaction between Menin and Mixed Lineage Leukemia (MLL).
J.Med.Chem., 59, 2016
|
|
2Y2B
 
 | | crystal structure of AmpD in complex with reaction products | | Descriptor: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD, 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, L-ALA-GAMMA-D-GLU-MESO-DIAMINOPIMELIC ACID, ... | | Authors: | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2010-12-14 | | Release date: | 2011-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
6M4L
 
 | | X-ray crystal structure of the E249Q mutant of alpha-amylase I from Eisenia fetida | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Hirano, Y, Tsukamoto, K, Ariki, S, Naka, Y, Ueda, M, Tamada, T. | | Deposit date: | 2020-03-07 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystallographic structural studies of alpha-amylase I from Eisenia fetida.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
4BXH
 
 | | Resolving the activation site of positive regulators in plant phosphoenolpyruvate carboxylase | | Descriptor: | 1,2-ETHANEDIOL, C4 PHOSPHOENOLPYRUVATE CARBOXYLASE, SULFATE ION | | Authors: | Schlieper, D, Foerster, K, Paulus, J.K, Groth, G. | | Deposit date: | 2013-07-11 | | Release date: | 2013-10-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Resolving the Activation Site of Positive Regulators in Plant Phosphoenolpyruvate Carboxylase.
Mol.Plant, 7, 2014
|
|
5T1A
 
 | | Structure of CC Chemokine Receptor 2 with Orthosteric and Allosteric Antagonists | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{R})-1-(4-chloranyl-2-fluoranyl-phenyl)-2-cyclohexyl-3-ethanoyl-4-oxidanyl-2~{H}-pyrrol-5-one, (3S)-1-{(1S,2R,4R)-4-[methyl(propan-2-yl)amino]-2-propylcyclohexyl}-3-{[6-(trifluoromethyl)quinazolin-4-yl]amino}pyrrolidin-2-one, ... | | Authors: | Zheng, Y, Qin, L, Ortiz Zacarias, N.V, de Vries, H, Han, G.W, Gustavsson, M, Dabros, M, Zhao, C, Cherney, R.J, Carter, P, Stamos, D, Abagyan, R, Cherezov, V, Stevens, R.C, IJzerman, A.P, Heitman, L.H, Tebben, A, Kufareva, I, Handel, T.M. | | Deposit date: | 2016-08-18 | | Release date: | 2016-12-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists.
Nature, 540, 2016
|
|
6F2U
 
 | | Potent and selective Aldo-Keto Reductase 1C3 (AKR1C3) inhibitors based on the benzoisoxazole moiety: application of a Bioisosteric Scaffold Hopping Approach to Flufenamic acid | | Descriptor: | 3-[(4-methoxyphenyl)methyl]-5-oxidanyl-~{N}-[3-(trifluoromethyl)phenyl]-1,2,3-triazole-4-carboxamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Goyal, P, Wahlgren, W.Y, Friemann, R. | | Deposit date: | 2017-11-27 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Potent and selective aldo-keto reductase 1C3 (AKR1C3) inhibitors based on the benzoisoxazole moiety: application of a bioisosteric scaffold hopping approach to flufenamic acid.
Eur J Med Chem, 150, 2018
|
|
5T1H
 
 | | Crystal structure of CK2 | | Descriptor: | 1,2-ETHANEDIOL, 7-(cyclopropylamino)-5-[3-(6-oxo-1,6-dihydropyridin-3-yl)thiophen-2-yl]pyrazolo[1,5-a]pyrimidine-3-carbonitrile, Casein kinase II subunit alpha, ... | | Authors: | Feguson, A.D. | | Deposit date: | 2016-08-19 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of CK2
To Be Published
|
|
3PHN
 
 | | Crystal structure of wild-type onconase with resolution 1.46 A | | Descriptor: | ACETATE ION, Protein P-30, SULFATE ION | | Authors: | Kurpiewska, K, Torrent, G, Ribo, M, Vilanova, M, Loch, J, Lewinski, K. | | Deposit date: | 2010-11-04 | | Release date: | 2010-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure of Rana pipiens wild-type onconase at resolution 1.46 A
To be Published
|
|
2VN4
 
 | | Glycoside Hydrolase Family 15 Glucoamylase from Hypocrea jecorina | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bott, R, Sandgren, M, Hansson, H. | | Deposit date: | 2008-01-30 | | Release date: | 2008-05-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Three-Dimensional Structure of an Intact Glycoside Hydrolase Family 15 Glucoamylase from Hypocrea Jecorina.
Biochemistry, 47, 2008
|
|
3GXK
 
 | | The crystal structure of g-type lysozyme from Atlantic cod (Gadus morhua L.) in complex with NAG oligomers sheds new light on substrate binding and the catalytic mechanism. Native structure to 1.9 | | Descriptor: | COBALT (II) ION, Goose-type lysozyme 1 | | Authors: | Helland, R, Larsen, R.L, Finstad, S, Kyomuhendo, P, Larsen, A.N. | | Deposit date: | 2009-04-02 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of g-type lysozyme from Atlantic cod shed new light on substrate binding and the catalytic mechanism.
Cell.Mol.Life Sci., 66, 2009
|
|
4NIR
 
 | |
2XFR
 
 | | Crystal structure of barley beta-amylase at atomic resolution | | Descriptor: | 1,2-ETHANEDIOL, BETA-AMYLASE | | Authors: | Rejzek, M, Stevenson, C.E.M, Southard, A.M, Stanley, D, Denyer, K, Smith, A.M, Naldrett, M.J, Lawson, D.M, Field, R.A. | | Deposit date: | 2010-05-28 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Chemical Genetics and Cereal Starch Metabolism: Structural Basis of the Non-Covalent and Covalent Inhibition of Barley Beta-Amylase.
Mol.Biosyst., 7, 2011
|
|
5UL5
 
 | | Crystal structure of RPE65 in complex with MB-004 and palmitate | | Descriptor: | (1R)-3-amino-1-{3-[(2-propylpentyl)oxy]phenyl}propan-1-ol, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, FE (II) ION, ... | | Authors: | Kiser, P.D, Palczewski, K. | | Deposit date: | 2017-01-24 | | Release date: | 2017-05-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational Tuning of Visual Cycle Modulator Pharmacodynamics.
J. Pharmacol. Exp. Ther., 362, 2017
|
|
5ULN
 
 | | Synthesis of novel seleno ureido containing compounds as SLC-0111 analogs. Investigations on carbonic anhydrases activity, glutathione peroxidase and X-ray crystallography | | Descriptor: | 4-{[(4-fluorophenyl)carbamothioyl]amino}benzene-1-sulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Peat, T.S, Angeli, A, Tanini, D, Bartolucci, G, Capperucci, A, Supuran, C.T, Carta, F. | | Deposit date: | 2017-01-25 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Discovery of New Selenoureido Analogues of 4-(4-Fluorophenylureido)benzenesulfonamide as Carbonic Anhydrase Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
5L5I
 
 | |
5L60
 
 | |
4C78
 
 | | Complex of human Sirt3 with Bromo-Resveratrol and ACS2 peptide | | Descriptor: | 5-[(E)-2-(4-bromophenyl)ethenyl]benzene-1,3-diol, ACETYL-COENZYME A SYNTHETASE 2-LIKE, MITOCHONDRIAL, ... | | Authors: | Nguyen, G.T.T, Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-09-19 | | Release date: | 2013-11-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Sirt3 Complexes with 4'-Bromo-Resveratrol Reveal Binding Sites and Inhibition Mechanism.
Chem.Biol., 20, 2013
|
|
5ZCD
 
 | | Crystal structure of Alpha-glucosidase in complex with maltotriose | | Descriptor: | Alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kato, K, Saburi, W, Yao, M. | | Deposit date: | 2018-02-16 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.707 Å) | | Cite: | Function and structure of GH13_31 alpha-glucosidase with high alpha-(1→4)-glucosidic linkage specificity and transglucosylation activity.
FEBS Lett., 592, 2018
|
|
6U7K
 
 | | Prefusion structure of PEDV spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wrapp, D, McLellan, J.S. | | Deposit date: | 2019-09-03 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | The 3.1-Angstrom Cryo-electron Microscopy Structure of the Porcine Epidemic Diarrhea Virus Spike Protein in the Prefusion Conformation.
J.Virol., 93, 2019
|
|
7DCF
 
 | | Crystal structure of EHMT2 SET domain in complex with compound 10 | | Descriptor: | 5'-methoxy-6'-(1-methyl-2,3,4,7-tetrahydroazepin-5-yl)spiro[cyclobutane-1,3'-indole]-2'-amine, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYLMETHIONINE, ... | | Authors: | Suzuki, M, Katayama, K. | | Deposit date: | 2020-10-26 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of DS79932728: A Potent, Orally Available G9a/GLP Inhibitor for Treating beta-Thalassemia and Sickle Cell Disease.
Acs Med.Chem.Lett., 12, 2021
|
|
3R0T
 
 | | Crystal structure of human protein kinase CK2 alpha subunit in complex with the inhibitor CX-5279 | | Descriptor: | 1,2-ETHANEDIOL, 3-(cyclopropylamino)-5-{[3-(trifluoromethyl)phenyl]amino}pyrimido[4,5-c]quinoline-8-carboxylic acid, Casein kinase II subunit alpha, ... | | Authors: | Battistutta, R, Papinutto, E, Lolli, G, Pierre, F, Haddach, M, Ryckman, D.M. | | Deposit date: | 2011-03-09 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Unprecedented selectivity and structural determinants of a new class of protein kinase CK2 inhibitors in clinical trials for the treatment of cancer.
Biochemistry, 50, 2011
|
|
4NHV
 
 | |
