1W5J
 
 | | AN ANTI-PARALLEL FOUR HELIX BUNDLE | | Descriptor: | GENERAL CONTROL PROTEIN GCN4 | | Authors: | Yadav, M.K, Leman, L.J, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2004-08-07 | | Release date: | 2004-09-24 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Coiled Coils at the Edge of Configurational Heterogeneity. Structural Analyses of Parallel and Antiparallel Homotetrameric Coiled Coils Reveal Configurational Sensitivity to a Single Solvent-Exposed Amino Acid Substitution.
Biochemistry, 45, 2006
|
|
2YLY
 
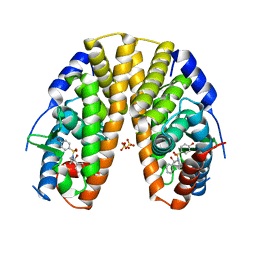 | | Sulfonamides as selective Estrogen Receptor beta Agonists. | | Descriptor: | ESTROGEN RECEPTOR BETA, N-cyclopropyl-4-oxidanyl-N-[(2R)-2-oxidanyl-2-phenyl-propyl]benzenesulfonamide, SULFATE ION | | Authors: | Roberts, L.R, Armour, D, Barker, C, Bazin, R, Bess, K, Brown, A, Favor, D, Ellis, D, MacKenny, M, Pullen, N, Stennett, A, Strand, L, Styles, M, Phillips, C. | | Deposit date: | 2011-06-06 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Sulfonamides as Selective Oestrogen Receptor Beta Agonists.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
5WXH
 
 | | Crystal structure of TAF3 PHD finger bound to H3K4me3 | | Descriptor: | Histone H3K4me3, Transcription initiation factor TFIID subunit 3, ZINC ION | | Authors: | Zhao, S, Huang, J, Li, H. | | Deposit date: | 2017-01-07 | | Release date: | 2017-08-16 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.297 Å) | | Cite: | Kinetic and high-throughput profiling of epigenetic interactions by 3D-carbene chip-based surface plasmon resonance imaging technology
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5WXG
 
 | | Structure of TAF PHD finger domain binds to H3(1-15)K4ac | | Descriptor: | Histone H3K4ac, MAGNESIUM ION, Transcription initiation factor TFIID subunit 3, ... | | Authors: | Zhao, S, Li, H. | | Deposit date: | 2017-01-07 | | Release date: | 2017-08-16 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Kinetic and high-throughput profiling of epigenetic interactions by 3D-carbene chip-based surface plasmon resonance imaging technology
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2A4D
 
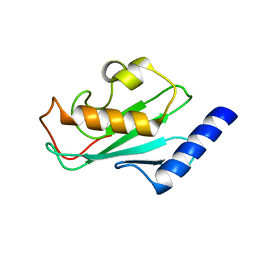 | | Structure of the human ubiquitin-conjugating enzyme E2 variant 1 (UEV-1) | | Descriptor: | Ubiquitin-conjugating enzyme E2 variant 1 | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Newman, E.M, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-28 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
Mol Cell Proteomics, 11, 2012
|
|
4IWC
 
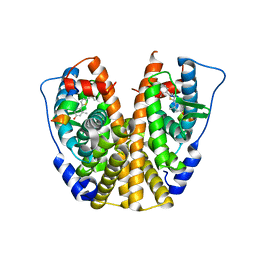 | | Crystal Structure of the Estrogen Receptor alpha Ligand-binding Domain in Complex with a Dynamic Thiophene-derivative | | Descriptor: | 4,4'-thiene-2,5-diylbis(3-methylphenol), Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Parent, A.A, Cavett, V, Nowak, J, Hughes, T.S, Kojetin, D.J, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2013-01-23 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Ligand binding dynamics rewire cellular signaling via Estrogen Receptor-alpha
Nat.Chem.Biol., 9, 2013
|
|
6UMP
 
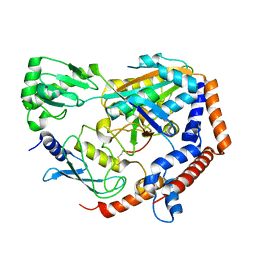 | | Crystal structure of MavC in complex with substrate mimic in P65 space group | | Descriptor: | MavC, Ubiquitin, Ubiquitin-conjugating enzyme E2 N | | Authors: | Puvar, K, Iyer, S, Luo, Z.Q, Das, C. | | Deposit date: | 2019-10-10 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Legionella effector MavC targets the Ube2N~Ub conjugate for noncanonical ubiquitination.
Nat Commun, 11, 2020
|
|
4IW6
 
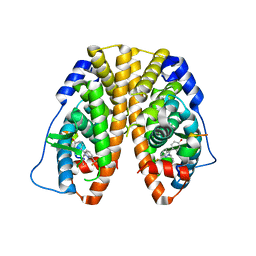 | | Crystal Structure of the Estrogen Receptor alpha Ligand-binding Domain in Complex with Constrained WAY-derivative, 7b | | Descriptor: | 4-[2-(but-3-en-1-yl)-7-(trifluoromethyl)-2H-indazol-3-yl]benzene-1,3-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Parent, A.A, Cavett, V, Nowak, J, Hughes, T.S, Kojetin, D.J, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2013-01-23 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Ligand binding dynamics rewire cellular signaling via Estrogen Receptor-alpha
Nat.Chem.Biol., 9, 2013
|
|
4IU7
 
 | | Crystal Structure of the Estrogen Receptor alpha Ligand-binding Domain in Complex with Constrained WAY-derivative, 2b | | Descriptor: | 4-[2-ethyl-7-(trifluoromethyl)-2H-indazol-3-yl]benzene-1,3-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Parent, A.A, Cavett, V, Nowak, J, Hughes, T.S, Kojetin, D.J, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2013-01-20 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Ligand binding dynamics rewire cellular signaling via Estrogen Receptor-alpha
Nat.Chem.Biol., 9, 2013
|
|
4IV4
 
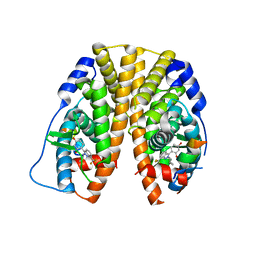 | | Crystal Structure of the Estrogen Receptor alpha Ligand-binding Domain in Complex with Constrained WAY-derivative, 5b | | Descriptor: | 4-[2-(2-methylpropyl)-7-(trifluoromethyl)-2H-indazol-3-yl]benzene-1,3-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Parent, A.A, Cavett, V, Nowak, J, Hughes, T.S, Kojetin, D.J, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2013-01-22 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ligand binding dynamics rewire cellular signaling via Estrogen Receptor-alpha
Nat.Chem.Biol., 9, 2013
|
|
4IVW
 
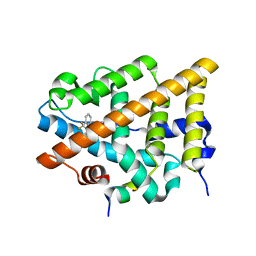 | | Crystal Structure of the Estrogen Receptor alpha Ligand-binding Domain in Complex with Constrained WAY-derivative, 6b | | Descriptor: | 4-[2-benzyl-7-(trifluoromethyl)-2H-indazol-3-yl]benzene-1,3-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Parent, A.A, Cavett, V, Nowak, J, Hughes, T.S, Kojetin, D.J, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2013-01-23 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Ligand binding dynamics rewire cellular signaling via Estrogen Receptor-alpha
Nat.Chem.Biol., 9, 2013
|
|
4IWF
 
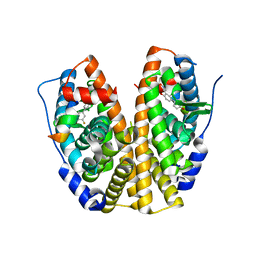 | | Crystal Structure of the Estrogen Receptor alpha Ligand-binding Domain in Complex with a Dynamic Oxime-derivative | | Descriptor: | 2-chloro-3'-fluoro-3-[(E)-(hydroxyimino)methyl]biphenyl-4,4'-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Parent, A.A, Cavett, V, Nowak, J, Hughes, T.S, Kojetin, D.J, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2013-01-23 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.932 Å) | | Cite: | Ligand binding dynamics rewire cellular signaling via Estrogen Receptor-alpha
Nat.Chem.Biol., 9, 2013
|
|
4IV2
 
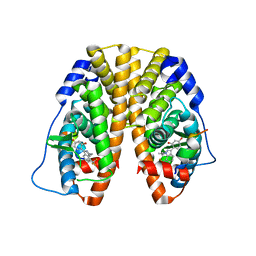 | | Crystal Structure of the Estrogen Receptor alpha Ligand-binding Domain in Complex with Dynamic WAY-derivative, 5a | | Descriptor: | 4-[1-(2-methylpropyl)-7-(trifluoromethyl)-1H-indazol-3-yl]benzene-1,3-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Parent, A.A, Cavett, V, Nowak, J, Hughes, T.S, Kojetin, D.J, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2013-01-22 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Ligand binding dynamics rewire cellular signaling via Estrogen Receptor-alpha
Nat.Chem.Biol., 9, 2013
|
|
4IW8
 
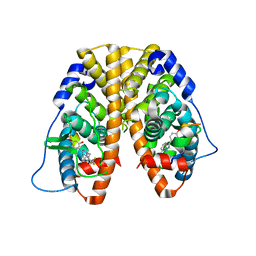 | | Crystal Structure of the Estrogen Receptor alpha Ligand-binding Domain in Complex with Dynamic WAY-derivative, 9a | | Descriptor: | 4-[1-(3-methylbut-2-en-1-yl)-7-(trifluoromethyl)-1H-indazol-3-yl]benzene-1,3-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Parent, A.A, Cavett, V, Nowak, J, Hughes, T.S, Kojetin, D.J, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2013-01-23 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.0375 Å) | | Cite: | Ligand binding dynamics rewire cellular signaling via Estrogen Receptor-alpha
Nat.Chem.Biol., 9, 2013
|
|
6KO7
 
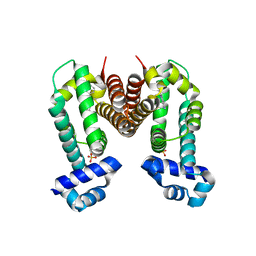 | | Crystal structure of the Ethidium bound RamR determined with XtaLAB Synergy | | Descriptor: | ETHIDIUM, Putative regulatory protein, SULFATE ION | | Authors: | Matsumoto, T, Nakashima, R, Yamano, A, Nishino, K. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of a structure determination method using a multidrug-resistance regulator protein as a framework.
Biochem.Biophys.Res.Commun., 518, 2019
|
|
4IUI
 
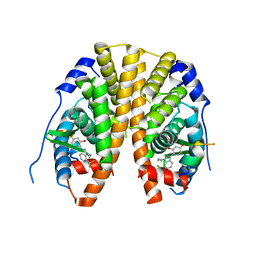 | | Crystal Structure of the Estrogen Receptor alpha Ligand-binding Domain in Complex with Dynamic WAY derivative, 4a | | Descriptor: | 4-[1-butyl-7-(trifluoromethyl)-1H-indazol-3-yl]benzene-1,3-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Parent, A.A, Cavett, V, Nowak, J, Hughes, T.S, Kojetin, D.J, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2013-01-21 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ligand binding dynamics rewire cellular signaling via Estrogen Receptor-alpha
Nat.Chem.Biol., 9, 2013
|
|
5XLP
 
 | |
1WH7
 
 | | Solution structure of homeobox domain of Arabidopsis thaliana hypothetical protein F22K18.140 | | Descriptor: | ZF-HD homeobox family protein | | Authors: | Kaneno, D, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of homeobox domain of Arabidopsis thaliana hypothetical protein F22K18.140
To be Published
|
|
6KO9
 
 | | Crystal structure of the Gefitinib Intermediate 1 bound RamR determined with XtaLAB Synergy | | Descriptor: | 4-[(3-chloranyl-4-fluoranyl-phenyl)amino]-7-methoxy-quinazolin-6-ol, Putative regulatory protein, SULFATE ION | | Authors: | Matsumoto, T, Nakashima, R, Yamano, A, Nishino, K. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Development of a structure determination method using a multidrug-resistance regulator protein as a framework.
Biochem.Biophys.Res.Commun., 518, 2019
|
|
6KO8
 
 | | Crystal structure of the Cholic acid bound RamR determined with XtaLAB Synergy | | Descriptor: | CHOLIC ACID, Putative regulatory protein, SULFATE ION | | Authors: | Matsumoto, T, Nakashima, R, Yamano, A, Nishino, K. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Development of a structure determination method using a multidrug-resistance regulator protein as a framework.
Biochem.Biophys.Res.Commun., 518, 2019
|
|
1J48
 
 | | Crystal Structure of Apo-C1027 | | Descriptor: | Apoprotein of C1027 | | Authors: | Chen, Y, Li, J, Liu, Y, Bartlam, M, Gao, Y, Jin, L, Tang, H, Shao, Y, Zhen, Y, Rao, Z. | | Deposit date: | 2001-07-30 | | Release date: | 2003-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Apo-C1027 and Computer Modeling Analysis of C1027 Chromophore- Protein Complex
To be published
|
|
5XLO
 
 | |
4AWX
 
 | | Moonlighting functions of FeoC in the regulation of ferrous iron transport in Feo | | Descriptor: | FERROUS IRON TRANSPORT PROTEIN B, FERROUS IRON TRANSPORT PROTEIN C, NICKEL (II) ION, ... | | Authors: | Hung, K.-W, Tsai, J.-Y, Juan, T.-H, Hsu, Y.-L, Hsiao, C.D, Huang, T.-H. | | Deposit date: | 2012-06-06 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Klebsiella Pneumoniae Nfeob/Feoc Complex and Roles of Feoc in Regulation of Fe2+ Transport by the Bacterial Feo System.
J.Bacteriol., 194, 2012
|
|
1UNY
 
 | | Structure Based Engineering of Internal Molecular Surfaces Of Four Helix Bundles | | Descriptor: | GENERAL CONTROL PROTEIN GCN4 | | Authors: | Yadav, M.K, Redman, J.E, Alvarez-Gutierrez, J.M, Zhang, Y, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2003-09-15 | | Release date: | 2004-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Engineering of Internal Cavities in Coiled-Coil Peptides
Biochemistry, 44, 2005
|
|
1UO3
 
 | | Structure Based Engineering of Internal Molecular Surfaces Of Four Helix Bundles | | Descriptor: | GENERAL CONTROL PROTEIN GCN4 | | Authors: | Yadav, M.K, Redman, J.E, Alvarez-Gutierrez, J.M, Zhang, Y, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2003-09-15 | | Release date: | 2004-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure-Based Engineering of Internal Cavities in Coiled-Coil Peptides
Biochemistry, 44, 2005
|
|
