5D3K
 
 | | Crystal structure of the thioesterase domain of deoxyerythronolide B synthase | | Descriptor: | CALCIUM ION, Erythronolide synthase, modules 5 and 6, ... | | Authors: | Bergeret, F, Argyropoulos, P, Boddy, C.N, Schmeing, T.M. | | Deposit date: | 2015-08-06 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Towards a characterization of the structural determinants of specificity in the macrocyclizing thioesterase for deoxyerythronolide B biosynthesis.
Biochim.Biophys.Acta, 1860, 2015
|
|
3GAH
 
 | | Structure of a F112H variant PduO-type ATP:corrinoid adenosyltransferase from Lactobacillus reuteri complexed with cobalamin and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, COBALAMIN, Cobalamin adenosyltransferase PduO-like protein, ... | | Authors: | St Maurice, M, Mera, P.E, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2009-02-17 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Residue Phe112 of the human-type corrinoid adenosyltransferase (PduO) enzyme of Lactobacillus reuteri is critical to the formation of the four-coordinate Co(II) corrinoid substrate and to the activity of the enzyme.
Biochemistry, 48, 2009
|
|
3ASK
 
 | | Structure of UHRF1 in complex with histone tail | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Histone H3.3, ZINC ION | | Authors: | Arita, K, Sugita, K, Unoki, M, Hamamoto, R, Sekiyama, N, Tochio, H, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2010-12-16 | | Release date: | 2012-01-25 | | Last modified: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (2.904 Å) | | Cite: | Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1MOW
 
 | | E-DreI | | Descriptor: | 5'-D(*CP*CP*AP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*AP*GP*TP*TP*CP*CP*GP*GP*CP*G)-3', 5'-D(*CP*GP*CP*CP*GP*GP*AP*AP*CP*TP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*TP*GP*G)-3', GLYCEROL, ... | | Authors: | Chevalier, B.S, Kortemme, T, Chadsey, M.S, Baker, D, Monnat Jr, R.J, Stoddard, B.L. | | Deposit date: | 2002-09-10 | | Release date: | 2002-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design, Activity and Structure of a Highly Specific Artificial Endonuclease
Mol.Cell, 10, 2002
|
|
3ASP
 
 | | Crystal structure of P domain from Norovirus Funabashi258 stain in the complex with A-antigen | | Descriptor: | Capsid protein, SODIUM ION, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kubota, T, Kumagai, A, Itoh, H, Furukawa, S, Narimatsu, H, Wakita, T, Ishii, K, Takeda, N, Someya, Y, Shirato, H. | | Deposit date: | 2010-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of Lewis antigens by genogroup I norovirus
J.Virol., 86, 2012
|
|
7RES
 
 | | HUMAN IMPDH1 TREATED WITH ATP, IMP, AND NAD+, OCTAMER-CENTERED | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, Isoform 5 of Inosine-5'-monophosphate dehydrogenase 1, ... | | Authors: | Burrell, A.L, Kollman, J.M. | | Deposit date: | 2021-07-13 | | Release date: | 2022-01-12 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | IMPDH1 retinal variants control filament architecture to tune allosteric regulation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5GW5
 
 | | Structure of TRiC-AMP-PNP | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Zang, Y, Jin, M, Wang, H, Cong, Y. | | Deposit date: | 2016-09-08 | | Release date: | 2016-10-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Staggered ATP binding mechanism of eukaryotic chaperonin TRiC (CCT) revealed through high-resolution cryo-EM.
Nat. Struct. Mol. Biol., 23, 2016
|
|
7RGL
 
 | | HUMAN RETINAL VARIANT IMPDH1(546) TREATED WITH ATP, IMP, NAD+, INTERFACE-CENTERED | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Burrell, A.L, Kollman, J.M. | | Deposit date: | 2021-07-15 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | IMPDH1 retinal variants control filament architecture to tune allosteric regulation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4FRA
 
 | | Crystal Structure of ABBA+UDP+Gal at pH 5.0 with MPD as the cryoprotectant | | Descriptor: | DI(HYDROXYETHYL)ETHER, Histo-blood group ABO system transferase, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Johal, A.R, Alfaro, J.A, Blackler, R.J, Schuman, B, Borisova, S.N, Evans, S.V. | | Deposit date: | 2012-06-26 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | pH-induced conformational changes in human ABO(H) blood group glycosyltransferases confirm the importance of electrostatic interactions in the formation of the semi-closed state.
Glycobiology, 24, 2014
|
|
2VLP
 
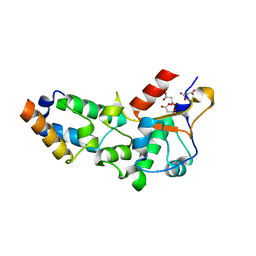 | | R54A mutant of E9 DNase domain in complex with Im9 | | Descriptor: | COLICIN E9, COLICIN-E9 IMMUNITY PROTEIN, MALONIC ACID | | Authors: | Keeble, A.H, Joachimiak, L.A, Mate, M.J, Meenan, N, Kirkpatrick, N, Baker, D, Kleanthous, C. | | Deposit date: | 2008-01-15 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Experimental and Computational Analyses of the Energetic Basis for Dual Recognition of Immunity Proteins by Colicin Endonucleases.
J.Mol.Biol., 379, 2008
|
|
7K3W
 
 | | Apoferritin structure at 1.36 angstrom resolution determined from a 300 kV Titan Krios G3i electron microscope with Falcon4 detector | | Descriptor: | Ferritin heavy chain, SODIUM ION, ZINC ION | | Authors: | Zhang, K, Pintilie, G, Li, S, Schmid, M, Chiu, W. | | Deposit date: | 2020-09-14 | | Release date: | 2020-11-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (1.36 Å) | | Cite: | Resolving individual atoms of protein complex by cryo-electron microscopy.
Cell Res., 30, 2020
|
|
3ATP
 
 | | Structure of the ligand binding domain of the bacterial serine chemoreceptor Tsr with ligand | | Descriptor: | Methyl-accepting chemotaxis protein I, SERINE | | Authors: | Tajima, H, Sakuma, M, Homma, K, Kawagishi, I, Imada, K. | | Deposit date: | 2011-01-07 | | Release date: | 2011-10-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand specificity determined by differentially arranged common ligand-binding residues in bacterial amino acid chemoreceptors Tsr and Tar.
J.Biol.Chem., 286, 2011
|
|
1MNB
 
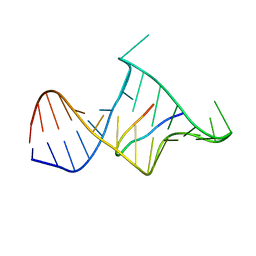 | | BIV TAT PEPTIDE (RESIDUES 68-81), NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | BIV TAR RNA, BIV TAT PEPTIDE | | Authors: | Puglisi, J.D, Chen, L, Blanchard, S, Frankel, A.D. | | Deposit date: | 1996-07-25 | | Release date: | 1997-01-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a bovine immunodeficiency virus Tat-TAR peptide-RNA complex.
Science, 270, 1995
|
|
5D6O
 
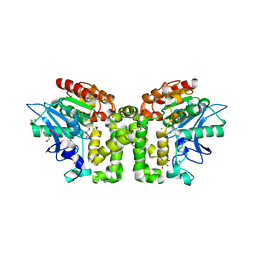 | | Orthorhombic Crystal Structure of an acetylester hydrolase from Corynebacterium glutamicum | | Descriptor: | CHLORIDE ION, GLYCEROL, Homoserine O-acetyltransferase, ... | | Authors: | Niefind, K, Toelzer, C, Altenbuchner, J, Watzlawick, H. | | Deposit date: | 2015-08-12 | | Release date: | 2015-12-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel esterase subfamily with alpha / beta-hydrolase fold suggested by structures of two bacterial enzymes homologous to l-homoserine O-acetyl transferases.
Febs Lett., 590, 2016
|
|
1MDI
 
 | |
7JQ1
 
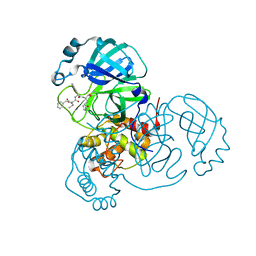 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI4 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl]propan-2-yl}-L-phenylalaninamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
5DET
 
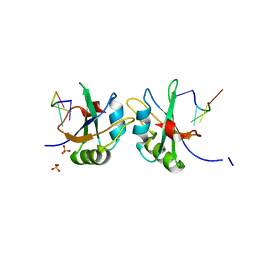 | | X-ray structure of human RBPMS in complex with the RNA | | Descriptor: | RNA (5'-R(*UP*CP*AP*C)-3'), RNA (5'-R(P*UP*CP*AP*CP*U)-3'), RNA-binding protein with multiple splicing, ... | | Authors: | Teplova, M, Farazi, T.A, Tuschl, T, Patel, D.J. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis underlying CAC RNA recognition by the RRM domain of dimeric RNA-binding protein RBPMS.
Q. Rev. Biophys., 49, 2016
|
|
2V0P
 
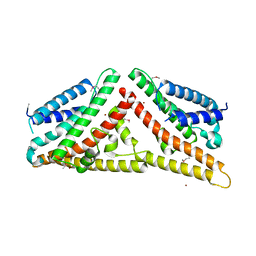 | | The Structure of Tap42 Alpha4 Subunit | | Descriptor: | TYPE 2A PHOSPHATASE-ASSOCIATED PROTEIN 42, ZINC ION | | Authors: | Roe, S.M, Yang, J, Barford, D. | | Deposit date: | 2007-05-15 | | Release date: | 2007-07-17 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of Tap42/Alpha4 Reveals a Tetratricopeptide Repeat-Like Fold and Provides Insights Into Pp2A Regulation.
Biochemistry, 46, 2007
|
|
7RFI
 
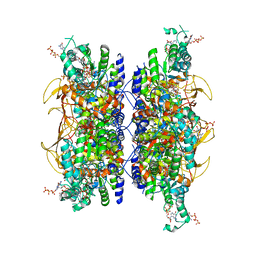 | | HUMAN RETINAL VARIANT IMPDH1(595) TREATED WITH GTP, ATP, IMP, NAD+, INTERFACE-CENTERED | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, Isoform 5 of Inosine-5'-monophosphate dehydrogenase 1, ... | | Authors: | Burrell, A.L, Kollman, J.M. | | Deposit date: | 2021-07-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | IMPDH1 retinal variants control filament architecture to tune allosteric regulation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
3GE7
 
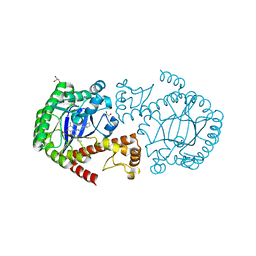 | | tRNA-guanine transglycosylase in complex with 6-amino-4-{2-[(cyclopentylmethyl)amino]ethyl}-2-(methylamino)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-amino-4-{2-[(cyclopentylmethyl)amino]ethyl}-2-(methylamino)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Ritschel, T, Heine, A, Klebe, G. | | Deposit date: | 2009-02-25 | | Release date: | 2009-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | How to Replace the Residual Solvation Shell of Polar Active Site Residues to Achieve Nanomolar Inhibition of tRNA-Guanine Transglycosylase
Chemmedchem, 4, 2009
|
|
8UFP
 
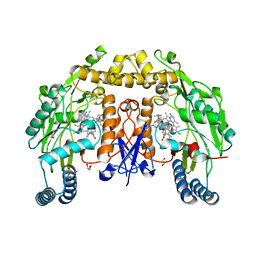 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 4-methyl-7-(4-methyl-2,3,4,5-tetrahydrobenzo[f][1,4]oxazepin-7-yl)quinolin-2-amine dihydrochloride | | Descriptor: | (7M)-4-methyl-7-(4-methyl-2,3,4,5-tetrahydro-1,4-benzoxazepin-7-yl)quinolin-2-amine, GLYCEROL, Nitric oxide synthase, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2023-10-04 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic and Computational Insights into Isoform-Selective Dynamics in Nitric Oxide Synthase.
Biochemistry, 63, 2024
|
|
5DF0
 
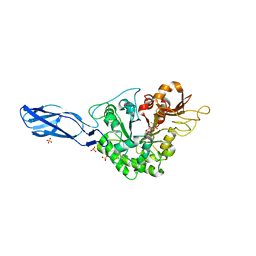 | | Crystal structure of AcMNPV Chitinase A in complex WITH CHITOTRIO-THIAZOLINE DITHIOAMIDE | | Descriptor: | (2R,3aR,5R,6R,7R,7aR)-5-(hydroxymethyl)-2-methylhexahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-2-(ethanethioylamino)-beta-D-glucopyranose, ... | | Authors: | Mou, T.-C, Sprang, S.R. | | Deposit date: | 2015-08-26 | | Release date: | 2016-09-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.251 Å) | | Cite: | Crystal structure of AcMNPV Chitinase A
To Be Published
|
|
7K7K
 
 | |
3G58
 
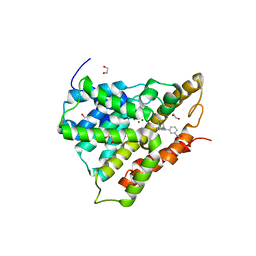 | | Crystal structure of human phosphodiesterase 4d with d155988/pmnpq | | Descriptor: | 1,2-ETHANEDIOL, 8-(3-nitrophenyl)-6-(pyridin-4-ylmethyl)quinoline, MAGNESIUM ION, ... | | Authors: | Staker, B.L. | | Deposit date: | 2009-02-04 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Design of phosphodiesterase 4D (PDE4D) allosteric modulators for enhancing cognition with improved safety.
Nat.Biotechnol., 28, 2010
|
|
4FL4
 
 | | Scaffoldin conformation and dynamics revealed by a ternary complex from the Clostridium thermocellum cellulosome | | Descriptor: | CALCIUM ION, Cellulosome anchoring protein cohesin region, Glycoside hydrolase family 9, ... | | Authors: | Currie, M.A, Adams, J.J, Faucher, F, Bayer, E.A, Jia, Z, Smith, S.P, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2012-06-14 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Scaffoldin Conformation and Dynamics Revealed by a Ternary Complex from the Clostridium thermocellum Cellulosome.
J.Biol.Chem., 287, 2012
|
|
