6TLX
 
 | | Crystal structure of the unconventional kinetochore protein Perkinsela sp. KKT2a central domain | | Descriptor: | Protein kinase, ZINC ION | | Authors: | Marciano, G, Nerusheva, O, Ishii, M, Akiyoshi, B. | | Deposit date: | 2019-12-03 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Kinetoplastid kinetochore proteins KKT2 and KKT3 have unique centromere localization domains.
J.Cell Biol., 220, 2021
|
|
7E0C
 
 | | Structure of L-glutamate oxidase R305E mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-glutamate oxidase | | Authors: | Ito, N, Matsuo, S, Inagaki, K, Imada, K. | | Deposit date: | 2021-01-27 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A new l-arginine oxidase engineered from l-glutamate oxidase.
Protein Sci., 30, 2021
|
|
6TMB
 
 | |
6TMM
 
 | | BIL2 domain from T.thermophila BUBL1 locus (C1A-N143A) | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Chiarini, V, Ilari, A. | | Deposit date: | 2019-12-04 | | Release date: | 2020-12-16 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Structural basis of ubiquitination mediated by protein splicing in early Eukarya.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
6T7P
 
 | | human plasmakallikrein protease domain in complex with active site directed inhibitor | | Descriptor: | (2~{S},4~{R})-1-[[(3~{S})-3-azanyl-2,3-dihydro-1-benzofuran-6-yl]carbonyl]-~{N}-(3-chlorophenyl)-4-phenyl-pyrrolidine-2-carboxamide, DIMETHYL SULFOXIDE, GLUTATHIONE, ... | | Authors: | Renatus, M. | | Deposit date: | 2019-10-22 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.416 Å) | | Cite: | Structure-Based Design and Preclinical Characterization of Selective and Orally Bioavailable Factor XIa Inhibitors: Demonstrating the Power of an Integrated S1 Protease Family Approach.
J.Med.Chem., 63, 2020
|
|
6TMS
 
 | | Crystal structure of a de novo designed hexameric helical-bundle protein | | Descriptor: | SULFATE ION, a novel designed pore protein, affinity purification tag | | Authors: | Xu, C, Pei, X.Y, Luisi, B.F, Baker, D. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Computational design of transmembrane pores.
Nature, 585, 2020
|
|
6TN0
 
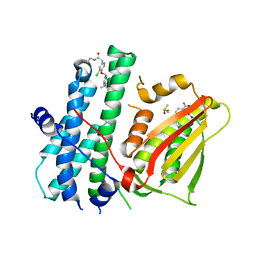 | | Rapid optimisation of fragments and hits to lead compounds from screening of crude reaction mixtures | | Descriptor: | 2,4-bis(oxidanyl)benzamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Baker, L.M, Aimon, A, Murray, J.B, Surgenor, A.E, Matassova, N, Roughley, S.D, von Delft, F, Hubbard, R.E. | | Deposit date: | 2019-12-05 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Rapid optimisation of fragments and hits to lead compounds from screening of crude reaction mixtures
Commun Chem, 2020
|
|
6TN4
 
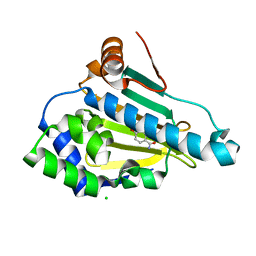 | | Rapid optimisation of fragments and hits to lead compounds from screening of crude reaction mixtures | | Descriptor: | 2,4-bis(oxidanyl)benzamide, CHLORIDE ION, Heat shock protein HSP 90-alpha | | Authors: | Baker, L.M, Aimon, A, Murray, J.B, Surgenor, A.E, Matassova, N, Roughley, S.D, von Delft, F, Hubbard, R.E. | | Deposit date: | 2019-12-05 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.274 Å) | | Cite: | Rapid optimisation of fragments and hits to lead compounds from screening of crude reaction mixtures
Commun Chem, 2020
|
|
7E7V
 
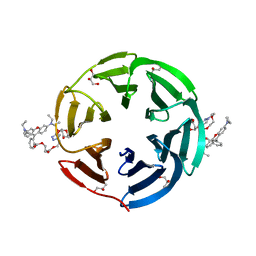 | | Crystal structure of RSL mutant in complex with sugar Ligand | | Descriptor: | 2-[2-[2-[4-[[(2R,3S,4R,5S,6S)-6-methyl-3,4,5-tris(oxidanyl)oxan-2-yl]oxymethyl]-1,2,3-triazol-1-yl]ethoxy]ethoxy]ethyl 2-[3,6-bis(diethylamino)-9H-xanthen-9-yl]benzoate, Fucose-binding lectin protein,Fucose-binding lectin protein,Fucose-binding lectin protein, GLYCEROL | | Authors: | Li, L, Chen, G.S. | | Deposit date: | 2021-02-28 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal structure of RSL mutant in complex with Ligand
To Be Published
|
|
6T80
 
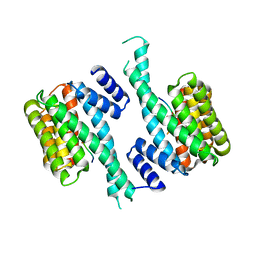 | |
7E7U
 
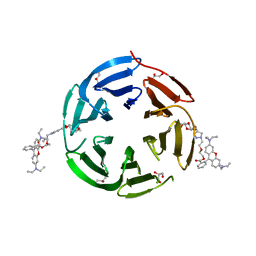 | | Crystal structure of RSL mutant in complex with sugar Ligand | | Descriptor: | 2-[2-[4-[[(2R,3S,4R,5S,6S)-6-methyl-3,4,5-tris(oxidanyl)oxan-2-yl]oxymethyl]-1,2,3-triazol-1-yl]ethoxy]ethyl 2-[3,6-bis(diethylamino)-9H-xanthen-9-yl]benzoate, Fucose-binding lectin protein,Fucose-binding lectin protein,Fucose-binding lectin protein, GLYCEROL | | Authors: | Li, L, Chen, G.S. | | Deposit date: | 2021-02-27 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of RSL mutant in complex with Ligand
To Be Published
|
|
6TO2
 
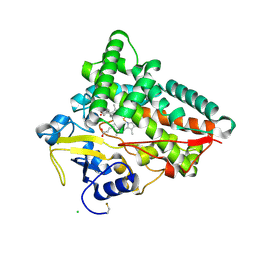 | | Crystal structure of CYP154C5 from Nocardia farcinica in complex with 5alpha-Androstan-3-one | | Descriptor: | (5~{S},8~{S},9~{S},10~{S},13~{S},14~{S})-10,13-dimethyl-1,2,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydrocyclopenta[a]phenanthren-3-one, CHLORIDE ION, Cytochrome P450 monooxygenase, ... | | Authors: | Rodriguez, A, Kluenemann, T, Blankenfeldt, W, Schallmey, A. | | Deposit date: | 2019-12-11 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CYP154C5 Regioselectivity in Steroid Hydroxylation Explored by Substrate Modifications and Protein Engineering*.
Chembiochem, 22, 2021
|
|
7E7R
 
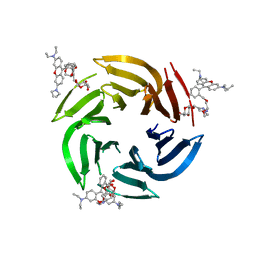 | | Crystal structure of RSL mutant in complex with Ligand | | Descriptor: | 2-[2-[4-[[(2R,3S,4R,5S,6S)-6-methyl-3,4,5-tris(oxidanyl)oxan-2-yl]oxymethyl]-1,2,3-triazol-1-yl]ethoxy]ethyl 2-[3,6-bis(diethylamino)-9H-xanthen-9-yl]benzoate, Fucose-binding lectin protein,Fucose-binding lectin protein,Fucose-binding lectin protein | | Authors: | Li, L, Chen, G.S. | | Deposit date: | 2021-02-27 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of RSL mutant-R17A/R108A/R199A in complex with R3F
To Be Published
|
|
6TP9
 
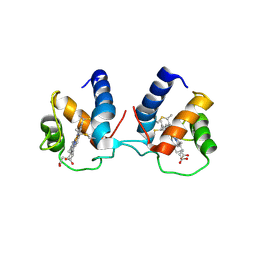 | | c-type cytochrome NirC | | Descriptor: | Cytochrome c55X, HEME C | | Authors: | Kluenemann, T, Henke, S, Blankenfeldt, W. | | Deposit date: | 2019-12-12 | | Release date: | 2020-04-22 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The crystal structure of the heme d1biosynthesis-associated small c-type cytochrome NirC reveals mixed oligomeric states in crystallo.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6T9R
 
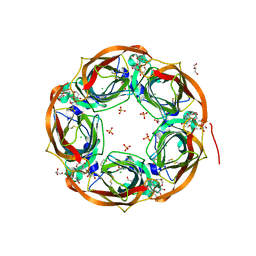 | | Aplysia californica AChBP in complex with a cytisine derivative | | Descriptor: | (1~{R},9~{S})-5-(3-oxidanylpropyl)-7,11-diazatricyclo[7.3.1.0^{2,7}]trideca-2,4-dien-6-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine binding protein, ... | | Authors: | Davis, S, Hunter, W.N. | | Deposit date: | 2019-10-28 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The thermodynamic profile and molecular interactions of a C(9)-cytisine derivative-binding acetylcholine-binding protein from Aplysia californica.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7E7N
 
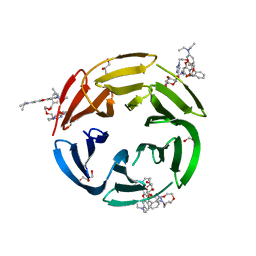 | | Crystal structure of RSL mutant-R17A/R108A/R199A in complex with R3F | | Descriptor: | 2-[2-[2-[4-[[(2R,3S,4R,5S,6S)-6-methyl-3,4,5-tris(oxidanyl)oxan-2-yl]oxymethyl]-1,2,3-triazol-1-yl]ethoxy]ethoxy]ethyl 2-[3,6-bis(diethylamino)-9H-xanthen-9-yl]benzoate, Fucose-binding lectin protein,Fucose-binding lectin protein,Fucose-binding lectin protein, GLYCEROL | | Authors: | Li, L, Chen, G.S. | | Deposit date: | 2021-02-26 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of RSL mutant in complex with suger Ligand
To Be Published
|
|
6TPY
 
 | |
6TRS
 
 | | Crystal structure of TFIIH subunit p52 in complex with p8 | | Descriptor: | RNA polymerase II transcription factor B subunit 2, Uncharacterized protein | | Authors: | Koelmel, W, Kuper, J, Schoenwetter, E, Kisker, C. | | Deposit date: | 2019-12-19 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | How to limit the speed of a motor: the intricate regulation of the XPB ATPase and translocase in TFIIH.
Nucleic Acids Res., 48, 2020
|
|
6TB1
 
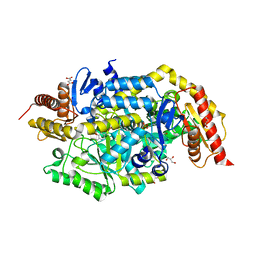 | | Crystal structure of thermostable omega transaminase 6-fold mutant from Pseudomonas jessenii | | Descriptor: | Aspartate aminotransferase family protein, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Capra, N, Rozeboom, H.J, Thunnissen, A.M.W.H, Janssen, D.B. | | Deposit date: | 2019-10-31 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Robust omega-Transaminases by Computational Stabilization of the Subunit Interface.
Acs Catalysis, 10, 2020
|
|
7ECC
 
 | | M4 family peptidase PlM4P-mature form | | Descriptor: | CALCIUM ION, M4 family peptidase, PHOSPHATE ION, ... | | Authors: | Yang, J. | | Deposit date: | 2021-03-12 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | M4 family peptidase PlM4P-mature form
To Be Published
|
|
6TB8
 
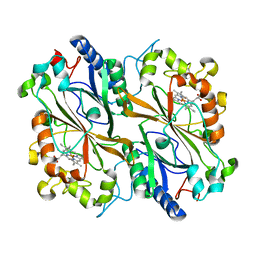 | | Dye Type Peroxidase Aa from Streptomyces lividans: spectroscopically-validated ferric state | | Descriptor: | Deferrochelatase/peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lucic, M, Moreno-Chicano, T.M, Hough, M.A, Dworkowski, F.S.N, Worrall, J.A.R. | | Deposit date: | 2019-11-01 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A subtle structural change in the distal haem pocket has a remarkable effect on tuning hydrogen peroxide reactivity in dye decolourising peroxidases from Streptomyces lividans.
Dalton Trans, 49, 2020
|
|
7E4L
 
 | |
6TBG
 
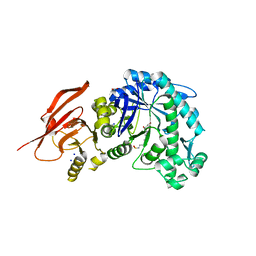 | | Structure of a beta galactosidase with inhibitor | | Descriptor: | 5-(dimethylamino)-~{N}-[6-[[(1~{R},2~{R},3~{S},4~{S},5~{S})-2-(hydroxymethyl)-3,4,5-tris(oxidanyl)cyclopentyl]amino]hexyl]naphthalene-1-sulfonamide, ACETATE ION, Beta-galactosidase, ... | | Authors: | Offen, W, Davies, G. | | Deposit date: | 2019-11-01 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanistic Insights into the Chaperoning of Human Lysosomal-Galactosidase Activity: Highly Functionalized Aminocyclopentanes and C -5a-Substituted Derivatives of 4- epi -Isofagomine.
Molecules, 25, 2020
|
|
7E9J
 
 | | Crystal Structure of POMGNT2 in complex with UDP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2, ... | | Authors: | Kuwabara, N. | | Deposit date: | 2021-03-04 | | Release date: | 2021-05-05 | | Last modified: | 2021-07-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of POMGNT2 provides new insights into the mechanism to determine the functional O-mannosylation site on alpha-dystroglycan.
Genes Cells, 26, 2021
|
|
6TC5
 
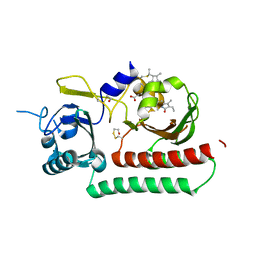 | | Phytochromobilin-adducted PAS-GAF bidomain of Sorghum bicolor phyB | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, BETA-MERCAPTOETHANOL, Phytochrome | | Authors: | Nagano, S, Guan, K, Shenkutie, S.M, Hughes, J.E. | | Deposit date: | 2019-11-05 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into photoactivation and signalling in plant phytochromes.
Nat.Plants, 6, 2020
|
|
