6VVC
 
 | |
6CHV
 
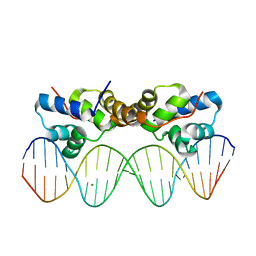 | | Proteus vulgaris HigA antitoxin bound to DNA | | Descriptor: | Antitoxin HigA, MAGNESIUM ION, pHigCryst3, ... | | Authors: | Schureck, M.A, Hoffer, E.D, Onuoha, N, Dunham, C.M. | | Deposit date: | 2018-02-23 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of transcriptional regulation by the HigA antitoxin.
Mol.Microbiol., 111, 2019
|
|
6PIC
 
 | |
3LSV
 
 | |
3IEU
 
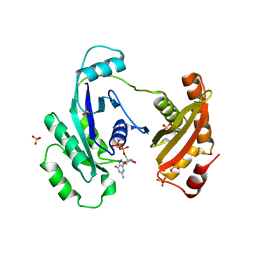 | | Crystal Structure of ERA in Complex with GDP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GTP-binding protein era, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Tu, C, Ji, X. | | Deposit date: | 2009-07-23 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of ERA in complex with the 3' end of 16S rRNA: implications for ribosome biogenesis
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6CZM
 
 | |
3LX2
 
 | |
3LZ0
 
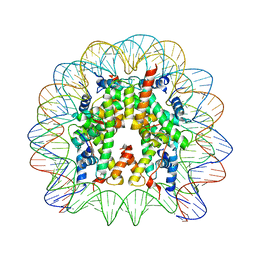 | | Crystal Structure of Nucleosome Core Particle Composed of the Widom 601 DNA Sequence (orientation 1) | | Descriptor: | CHLORIDE ION, DNA (145-MER), Histone H2A, ... | | Authors: | Vasudevan, D, Chua, E.Y.D, Davey, C.A. | | Deposit date: | 2010-03-01 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of nucleosome core particles containing the '601' strong positioning sequence
J.Mol.Biol., 403, 2010
|
|
3LZ1
 
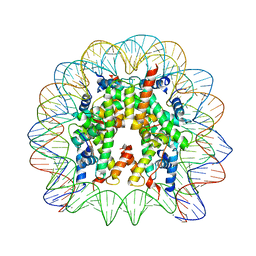 | | Crystal Structure of Nucleosome Core Particle Composed of the Widom 601 DNA Sequence (orientation 2) | | Descriptor: | CHLORIDE ION, DNA (145-MER), Histone H2A, ... | | Authors: | Vasudevan, D, Chua, E.Y.D, Davey, C.A. | | Deposit date: | 2010-03-01 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of nucleosome core particles containing the '601' strong positioning sequence
J.Mol.Biol., 403, 2010
|
|
3LX1
 
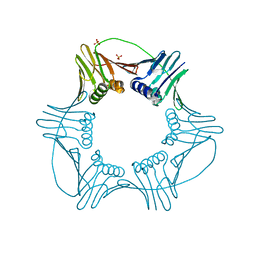 | |
3M1J
 
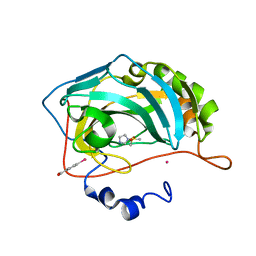 | |
3MCY
 
 | |
3IEV
 
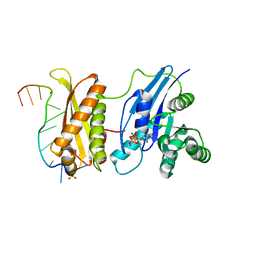 | | Crystal Structure of ERA in Complex with MgGNP and the 3' End of 16S rRNA | | Descriptor: | 5'-R(P*AP*UP*CP*AP*CP*CP*UP*CP*CP*UP*UP*A)-3', GTP-binding protein era, MAGNESIUM ION, ... | | Authors: | Tu, C, Ji, X. | | Deposit date: | 2009-07-23 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of ERA in complex with the 3' end of 16S rRNA: implications for ribosome biogenesis
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3MGR
 
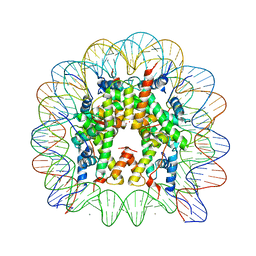 | | Binding of Rubidium ions to the Nucleosome Core Particle | | Descriptor: | CHLORIDE ION, DNA (147-MER), Histone H2A, ... | | Authors: | Mohideen, K, Muhammad, R, Davey, C.A. | | Deposit date: | 2010-04-07 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Perturbations in nucleosome structure from heavy metal association.
Nucleic Acids Res., 38, 2010
|
|
4E4W
 
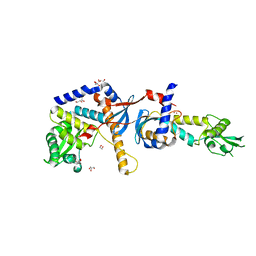 | | Structure of the C-terminal domain of the Saccharomyces cerevisiae MUTL alpha (MLH1/PMS1) heterodimer | | Descriptor: | 1,2-ETHANEDIOL, DNA mismatch repair protein MLH1, DNA mismatch repair protein PMS1, ... | | Authors: | Gueneau, E, Legrand, P, Charbonnier, J.B. | | Deposit date: | 2012-03-13 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the MutLalpha C-terminal domain reveals how Mlh1 contributes to Pms1 endonuclease site.
Nat.Struct.Mol.Biol., 20, 2013
|
|
6O58
 
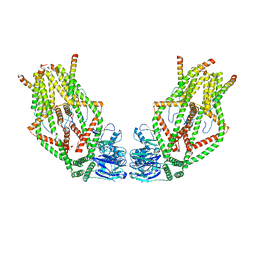 | | Human MCU-EMRE complex, dimer of channel | | Descriptor: | CALCIUM ION, Calcium uniporter protein, mitochondrial, ... | | Authors: | Wang, Y, Bai, X, Jiang, Y. | | Deposit date: | 2019-03-01 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural Mechanism of EMRE-Dependent Gating of the Human Mitochondrial Calcium Uniporter.
Cell, 177, 2019
|
|
6O5B
 
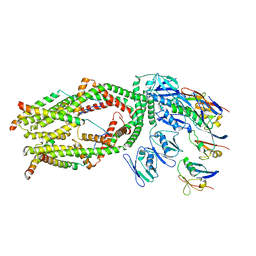 | | Monomer of a cation channel | | Descriptor: | CALCIUM ION, Calcium uniporter protein, mitochondrial, ... | | Authors: | Wang, Y, Bai, X, Jiang, Y. | | Deposit date: | 2019-03-01 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Mechanism of EMRE-Dependent Gating of the Human Mitochondrial Calcium Uniporter.
Cell, 177, 2019
|
|
6BHP
 
 | |
1WAY
 
 | | Active site thrombin inhibitors | | Descriptor: | 4-[3-(4-CHLOROPHENYL)-1H-PYRAZOL-5-YL]PIPERIDINE, DIMETHYL SULFOXIDE, HIRUGEN, ... | | Authors: | Hartshorn, M.J, Murray, C.W, Cleasby, A, Frederickson, M, Tickle, I.J, Jhoti, H. | | Deposit date: | 2004-10-28 | | Release date: | 2005-01-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Fragment-Based Lead Discovery Using X-Ray Crystallography
J.Med.Chem., 48, 2005
|
|
1WBG
 
 | | Active site thrombin inhibitors | | Descriptor: | 3-(4-CHLOROPHENYL)-5-(METHYLTHIO)-4H-1,2,4-TRIAZOLE, DIMETHYL SULFOXIDE, HIRUGEN, ... | | Authors: | Hartshorn, M.J, Murray, C.W, Cleasby, A, Frederickson, M, Tickle, I.J, Jhoti, H. | | Deposit date: | 2004-11-01 | | Release date: | 2005-01-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fragment-Based Lead Discovery Using X-Ray Crystallography
J.Med.Chem., 48, 2005
|
|
3H4C
 
 | | Structure of the C-terminal Domain of Transcription Factor IIB from Trypanosoma brucei | | Descriptor: | 1,2-ETHANEDIOL, Transcription factor TFIIB-like | | Authors: | Syed Ibrahim, B, Kanneganti, N, Rieckhof, G.E, Das, A, Laurents, D.V, Palenchar, J.B, Bellofatto, V, Wah, D.A. | | Deposit date: | 2009-04-18 | | Release date: | 2009-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the C-terminal domain of transcription factor IIB from Trypanosoma brucei.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8Y0X
 
 | | Dormant ribosome with SERBP1 | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Du, M, Zeng, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-02-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Implication of Stm1 in the protection of eIF5A, eEF2 and tRNA through dormant ribosomes.
Front Mol Biosci, 11, 2024
|
|
4FMN
 
 | | Structure of the C-terminal domain of the Saccharomyces cerevisiae MUTL alpha (MLH1/PMS1) heterodimer bound to a fragment of NTG2 | | Descriptor: | 1,2-ETHANEDIOL, DNA mismatch repair protein MLH1, DNA mismatch repair protein PMS1, ... | | Authors: | Gueneau, E, Legrand, P, Charbonnier, J.B. | | Deposit date: | 2012-06-18 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure of the MutL alpha C-terminal domain reveals how Mlh1 contributes to Pms1 endonuclease site.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4FMO
 
 | | Structure of the C-terminal domain of the Saccharomyces cerevisiae MUTL alpha (MLH1/PMS1) heterodimer bound to a fragment of exo1 | | Descriptor: | DNA mismatch repair protein MLH1, DNA mismatch repair protein PMS1, DNA repair peptide, ... | | Authors: | Gueneau, E, Legrand, P, Charbonnier, J.B. | | Deposit date: | 2012-06-18 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structure of the MutL alpha C-terminal domain reveals how Mlh1 contributes to Pms1 endonuclease site.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4FU5
 
 | |
