8JLO
 
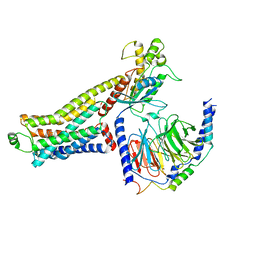 | | Ulotaront(SEP-363856)-bound hTAAR1-Gs protein complex | | Descriptor: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-15 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8JLR
 
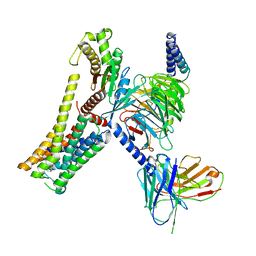 | | A77636-bound hTAAR1-Gs protein complex | | Descriptor: | (1~{S},3~{R})-3-(1-adamantyl)-1-(aminomethyl)-3,4-dihydro-1~{H}-isochromene-5,6-diol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-15 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8JLJ
 
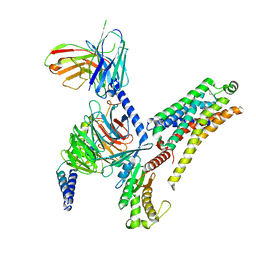 | | T1AM-bound mTAAR1-Gs protein complex | | Descriptor: | 4-[4-(2-azanylethyl)-2-iodanyl-phenoxy]phenol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8JSO
 
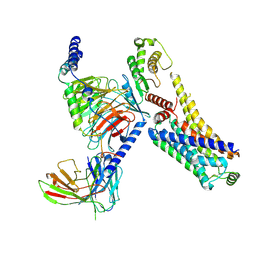 | | AMPH-bound hTAAR1-Gs protein complex | | Descriptor: | (2S)-1-phenylpropan-2-amine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
9DNT
 
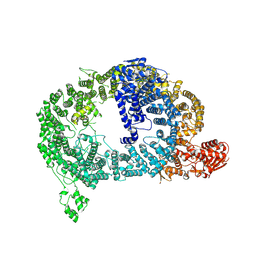 | | Cryo-EM structure of Tom1 (S. cerevisiae) | | Descriptor: | E3 ubiquitin-protein ligase TOM1 | | Authors: | Warner, K.M, Hunkeler, M, Baek, K, Roy Burman, S.S, Fischer, E.S. | | Deposit date: | 2024-09-18 | | Release date: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural ubiquitin contributes to K48 linkage specificity of the HECT ligase Tom1.
Cell Rep, 44, 2025
|
|
6HE2
 
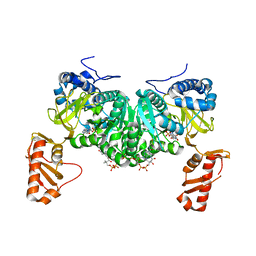 | | Crystal structure of an open conformation of 2-Hydroxyisobutyryl-CoA Ligase (HCL) in complex with 2-HIB-AMP and CoA | | Descriptor: | 2-hydroxyisobutyryl-CoA synthetase, ADENOSINE MONOPHOSPHATE, COENZYME A, ... | | Authors: | Zahn, M, Rohwerder, T, Strater, N. | | Deposit date: | 2018-08-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of 2-Hydroxyisobutyric Acid-CoA Ligase Reveal Determinants of Substrate Specificity and Describe a Multi-Conformational Catalytic Cycle.
J.Mol.Biol., 431, 2019
|
|
6HDX
 
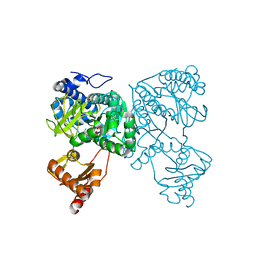 | | Crystal structure of 2-Hydroxyisobutyryl-CoA Ligase (HCL) in the postadenylation state in complex with R3-HIB-AMP | | Descriptor: | (2R)-3-HYDROXY-2-METHYLPROPANOIC ACID, 2-hydroxyisobutyryl-CoA synthetase, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] (2~{R})-2-methyl-3-oxidanyl-propanoate | | Authors: | Zahn, M, Rohwerder, T, Strater, N. | | Deposit date: | 2018-08-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of 2-Hydroxyisobutyric Acid-CoA Ligase Reveal Determinants of Substrate Specificity and Describe a Multi-Conformational Catalytic Cycle.
J.Mol.Biol., 431, 2019
|
|
1XJ3
 
 | | bjFixLH in unliganded ferrous form | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Sensor protein fixL | | Authors: | Key, J, Moffat, K. | | Deposit date: | 2004-09-22 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Deoxy and CO-Bound bjFixLH Reveal Details of Ligand Recognition and Signaling
Biochemistry, 44, 2005
|
|
4CA1
 
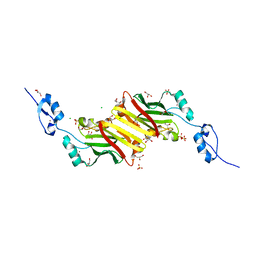 | | Crystal structure of Siah1 at 1.58 A resolution. | | Descriptor: | CHLORIDE ION, E3 UBIQUITIN-PROTEIN LIGASE SIAH1, GLYCEROL, ... | | Authors: | Rimsa, V, Eadsforth, T.C, Hunter, W.N. | | Deposit date: | 2013-10-04 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Two High-Resolution Structures of the Human E3 Ubiquitin Ligase Siah1.
Acta Crystallogr.,Sect.F, 96, 2013
|
|
1XJ6
 
 | | Structure of bjFixLH in the unliganded ferrous form | | Descriptor: | CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Key, J, Moffat, K. | | Deposit date: | 2004-09-22 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Deoxy and CO-Bound bjFixLH Reveal Details of Ligand Recognition and Signaling
Biochemistry, 44, 2005
|
|
2Z1P
 
 | |
6TYZ
 
 | |
6TYV
 
 | |
6Z4R
 
 | |
6Z4T
 
 | |
6TYX
 
 | |
1M5E
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH ACPA AT 1.46 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(3-CARBOXY-5-METHYLISOXAZOL-4-YL)PROPIONIC ACID, ACETATE ION, Glutamate receptor 2, ... | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
6TYU
 
 | |
3SGI
 
 | |
8PWL
 
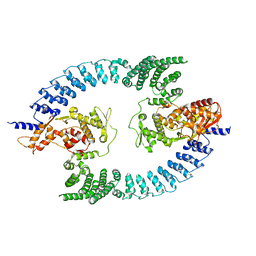 | |
6HC9
 
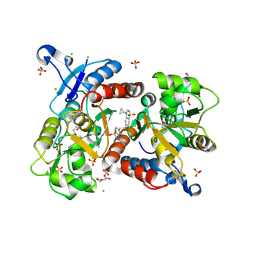 | | STRUCTURE OF GLUA2 LIGAND-BINDING DOMAIN (S1S2J-L504Y-N775S) IN COMPLEX WITH GLUTAMATE AND TDPAM02 AT 2.4 A RESOLUTION. | | Descriptor: | 6,6'-(ETHANE-1,2-DIYL)BIS(4-CYCLOPROPYL-3,4-DIHYDRO-2H-1,2,4-BENZOTHIADIAZINE 1,1-DIOXIDE), CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Laulumaa, S, Hansen, K.V, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2018-08-14 | | Release date: | 2019-04-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Potent Dimeric Positive Allosteric Modulators at the Ligand-Binding Domain of the GluA2 Receptor.
Acs Med.Chem.Lett., 10, 2019
|
|
6HCH
 
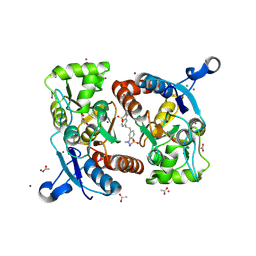 | | STRUCTURE OF GLUA2 LIGAND-BINDING DOMAIN (S1S2J-L504Y-N775S) IN COMPLEX WITH GLUTAMATE AND TDPAM01 AT 1.6 A RESOLUTION. | | Descriptor: | 6,6'-(Ethane-1,2-diyl)bis(4-methyl-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide), ACETATE ION, CHLORIDE ION, ... | | Authors: | Laulumaa, S, Hansen, K.V, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2018-08-15 | | Release date: | 2019-04-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Potent Dimeric Positive Allosteric Modulators at the Ligand-Binding Domain of the GluA2 Receptor.
Acs Med.Chem.Lett., 10, 2019
|
|
6WGN
 
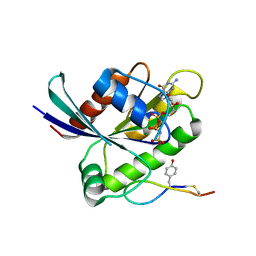 | | Crystal structure of K-Ras(G12D) GppNHp bound to cyclic peptide ligand KD2 | | Descriptor: | Cyclic Peptide KD2, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Zhang, Z, Gao, R, Hu, Q, Peacock, H, Peacock, D.M, Shokat, K.M, Suga, H. | | Deposit date: | 2020-04-06 | | Release date: | 2020-10-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | GTP-State-Selective Cyclic Peptide Ligands of K-Ras(G12D) Block Its Interaction with Raf.
Acs Cent.Sci., 6, 2020
|
|
6TYW
 
 | |
1AKY
 
 | |
