1B6M
 
 | | HIV-1 PROTEASE COMPLEXED WITH MACROCYCLIC PEPTIDOMIMETIC INHIBITOR 6 | | Descriptor: | RETROPEPSIN, SULFATE ION, [1-BENZYL-3-(8-SEC-BUTYL-7,10-DIOXO-2-OXA-6,9-DIAZA-BICYCLO[11.2.2] HEPTADECA-1(16),13(17),14-TRIEN-11-YLAMINO)-2-HYDROXY-PROPYL]-CARBAMIC ACID TERT-BUTYL ESTER | | Authors: | Martin, J.L, Begun, J, Schindeler, A, Wickramasinghe, W.A, Alewood, D, Alewood, P.F, Bergman, D.A, Brinkworth, R.I, Abbenante, G, March, D.R, Reid, R.C, Fairlie, D.P. | | Deposit date: | 1999-01-17 | | Release date: | 2000-01-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular recognition of macrocyclic peptidomimetic inhibitors by HIV-1 protease.
Biochemistry, 38, 1999
|
|
1JKH
 
 | | CRYSTAL STRUCTURE OF Y181C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH DMP-266(EFAVIRENZ) | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, HIV-1 RT, A-CHAIN, ... | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-12 | | Release date: | 2001-10-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
2F48
 
 | |
1AJK
 
 | | CIRCULARLY PERMUTED (1-3,1-4)-BETA-D-GLUCAN 4-GLUCANOHYDROLASE CPA16M-84 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CIRCULARLY PERMUTED (1-3,1-4)-BETA-D-GLUCAN 4-GLUCANOHYDROLASE, ... | | Authors: | Ay, J, Heinemann, U. | | Deposit date: | 1997-05-06 | | Release date: | 1998-05-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures and properties of de novo circularly permuted 1,3-1,4-beta-glucanases.
Proteins, 30, 1998
|
|
1TVR
 
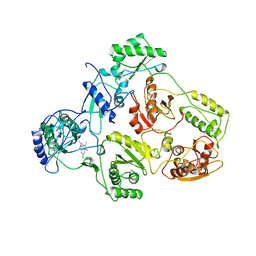 | | HIV-1 RT/9-CL TIBO | | Descriptor: | 4-CHLORO-8-METHYL-7-(3-METHYL-BUT-2-ENYL)-6,7,8,9-TETRAHYDRO-2H-2,7,9A-TRIAZA-BENZO[CD]AZULENE-1-THIONE, REVERSE TRANSCRIPTASE | | Authors: | Das, K, Ding, J, Hsiou, Y, Arnold, E. | | Deposit date: | 1996-04-16 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of 8-Cl and 9-Cl TIBO complexed with wild-type HIV-1 RT and 8-Cl TIBO complexed with the Tyr181Cys HIV-1 RT drug-resistant mutant.
J.Mol.Biol., 264, 1996
|
|
1B6K
 
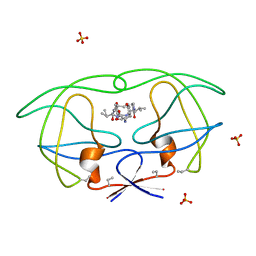 | | HIV-1 PROTEASE COMPLEXED WITH MACROCYCLIC PEPTIDOMIMETIC INHIBITOR 5 | | Descriptor: | N-[3-(8-SEC-BUTYL-7,10-DIOXO-2-OXA-6,9-DIAZA-BICYCLO[11.2.2]HEPTADECA-1(16),13(17),14- TRIEN-11-YLAMINO)-2-HYDROXY-1-(4-HYDROXY-BENZYL)-PROPYL]-3-METHYL-2- (2-OXO-PYRROLIDIN-1-YL)-BUTYRAMIDE, RETROPEPSIN, SULFATE ION | | Authors: | Martin, J.L, Begun, J, Schindeler, A, Wickramasinghe, W.A, Alewood, D, Alewood, P.F, Bergman, D.A, Brinkworth, R.I, Abbenante, G, March, D.R, Reid, R.C, Fairlie, D.P. | | Deposit date: | 1999-01-17 | | Release date: | 2000-01-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular recognition of macrocyclic peptidomimetic inhibitors by HIV-1 protease.
Biochemistry, 38, 1999
|
|
2GLX
 
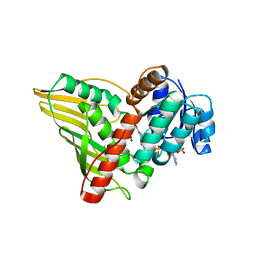 | | Crystal Structure Analysis of bacterial 1,5-AF Reductase | | Descriptor: | 1,5-anhydro-D-fructose reductase, ACETATE ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Dambe, T.R, Scheidig, A.J. | | Deposit date: | 2006-04-05 | | Release date: | 2006-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of NADP(H)-Dependent 1,5-Anhydro-d-fructose Reductase from Sinorhizobium morelense at 2.2 A Resolution: Construction of a NADH-Accepting Mutant and Its Application in Rare Sugar Synthesis
Biochemistry, 45, 2006
|
|
2HRQ
 
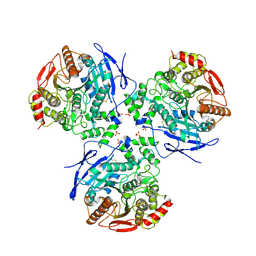 | | Crystal structure of Human Liver Carboxylesterase 1 (hCE1) in covalent complex with the nerve agent Soman (GD) | | Descriptor: | (1R)-1,2,2-TRIMETHYLPROPYL (R)-METHYLPHOSPHINATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Liver carboxylesterase 1, ... | | Authors: | Fleming, C.D, Redinbo, M.R. | | Deposit date: | 2006-07-20 | | Release date: | 2007-05-01 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of Human Carboxylesterase 1 in Covalent Complexes with the Chemical Warfare Agents Soman and Tabun.
Biochemistry, 46, 2007
|
|
2JCV
 
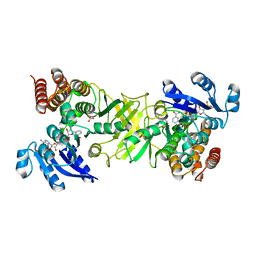 | | X-ray structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase, DXR, Rv2870c, from Mycobacterium tuberculosis, in complex with fosmidomycin and NADPH | | Descriptor: | 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Henriksson, L.M, Unge, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2007-01-04 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of Mycobacterium Tuberculosis 1-Deoxy-D- Xylulose-5-Phosphate Reductoisomerase Provide New Insights Into Catalysis.
J.Biol.Chem., 282, 2007
|
|
2JD2
 
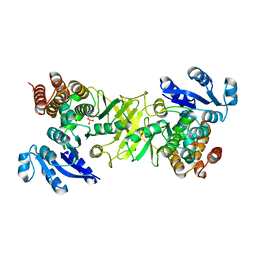 | | X-ray structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase, DXR, Rv2870c, from Mycobacterium tuberculosis, in complex with manganese | | Descriptor: | 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE, MANGANESE (II) ION, SULFATE ION | | Authors: | Henriksson, L.M, Unge, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2007-01-04 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of Mycobacterium Tuberculosis 1-Deoxy-D- Xylulose-5-Phosphate Reductoisomerase Provide New Insights Into Catalysis.
J.Biol.Chem., 282, 2007
|
|
2JD0
 
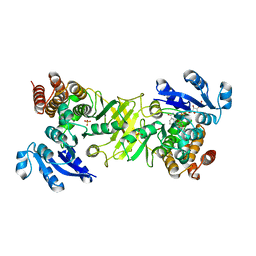 | | X-ray structure of mutant 1-deoxy-D-xylulose 5-phosphate reductoisomerase, DXR, Rv2870c, from Mycobacterium tuberculosis, in complex with NADPH | | Descriptor: | 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Henriksson, L.M, Unge, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2007-01-04 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Mycobacterium Tuberculosis 1-Deoxy-D- Xylulose-5-Phosphate Reductoisomerase Provide New Insights Into Catalysis.
J.Biol.Chem., 282, 2007
|
|
2IY6
 
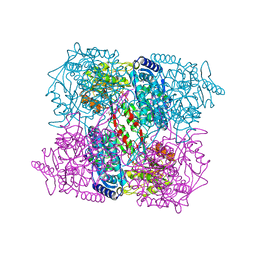 | | 1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE FROM THERMUS WITH BOUND CITRATE | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE, ... | | Authors: | Inagaki, E, Sakamoto, K, Nishio, M, Yokoyama, S. | | Deposit date: | 2006-07-13 | | Release date: | 2006-07-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Thermus Thermophilus Delta(1)- Pyrroline-5-Carboxylate Dehydrogenase.
J.Mol.Biol., 362, 2006
|
|
2JCY
 
 | | X-ray structure of mutant 1-deoxy-D-xylulose 5-phosphate reductoisomerase, DXR, Rv2870c, from Mycobacterium tuberculosis | | Descriptor: | 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE, SULFATE ION | | Authors: | Henriksson, L.M, Unge, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2007-01-04 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of Mycobacterium Tuberculosis 1-Deoxy-D- Xylulose-5-Phosphate Reductoisomerase Provide New Insights Into Catalysis.
J.Biol.Chem., 282, 2007
|
|
2PE4
 
 | | Structure of Human Hyaluronidase 1, a Hyaluronan Hydrolyzing Enzyme Involved in Tumor Growth and Angiogenesis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, GLYCEROL, ... | | Authors: | Chao, K.L, Herzberg, O. | | Deposit date: | 2007-04-02 | | Release date: | 2007-06-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Human Hyaluronidase-1, a Hyaluronan Hydrolyzing Enzyme Involved in Tumor Growth and Angiogenesis
Biochemistry, 46, 2007
|
|
2JD1
 
 | | X-ray structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase, DXR, Rv2870c, from Mycobacterium tuberculosis, in complex with manganese and NADPH | | Descriptor: | 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE, MANGANESE (II) ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Henriksson, L.M, Unge, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2007-01-04 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Mycobacterium Tuberculosis 1-Deoxy-D- Xylulose-5-Phosphate Reductoisomerase Provide New Insights Into Catalysis.
J.Biol.Chem., 282, 2007
|
|
1S1X
 
 | | Crystal structure of V108I mutant HIV-1 reverse transcriptase in complex with nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, Reverse transcriptase | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
1S1V
 
 | | Crystal structure of L100I mutant HIV-1 reverse transcriptase in complex with TNK-651 | | Descriptor: | 6-BENZYL-1-BENZYLOXYMETHYL-5-ISOPROPYL URACIL, Reverse transcriptase | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
1S1U
 
 | | Crystal structure of L100I mutant HIV-1 reverse transcriptase in complex with nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, Reverse transcriptase | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
2CJT
 
 | | Structural Basis for a Munc13-1 Homodimer - Munc13-1 - RIM Heterodimer Switch: C2-domains as Versatile Protein-Protein Interaction Modules | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, UNC-13 HOMOLOG A | | Authors: | Lu, J, Machius, M, Dulubova, I, Dai, H, Sudhof, T.C, Tomchick, D.R, Rizo, J. | | Deposit date: | 2006-04-06 | | Release date: | 2006-06-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural Basis for a Munc13-1 Dimeric to Munc13-1/Rim Heterodimer Switch
Plos Biol., 4, 2006
|
|
2G0A
 
 | | X-ray structure of mouse pyrimidine 5'-nucleotidase type 1 with lead(II) bound in active site | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cytosolic 5'-nucleotidase III, LEAD (II) ION | | Authors: | Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-02-11 | | Release date: | 2006-04-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of pyrimidine 5'-nucleotidase type 1. Insight into mechanism of action and inhibition during lead poisoning.
J.Biol.Chem., 281, 2006
|
|
2JCX
 
 | | X-ray structure of mutant 1-deoxy-D-xylulose 5-phosphate reductoisomerase, DXR, Rv2870c, from Mycobacterium tuberculosis, in complex with fosmidomycin and NADPH | | Descriptor: | 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Henriksson, L.M, Unge, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2007-01-04 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Mycobacterium Tuberculosis 1-Deoxy-D- Xylulose-5-Phosphate Reductoisomerase Provide New Insights Into Catalysis.
J.Biol.Chem., 282, 2007
|
|
2M3Z
 
 | |
2RI9
 
 | | Penicillium citrinum alpha-1,2-mannosidase in complex with a substrate analog | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lobsanov, Y.D, Yoshida, T, Desmet, T, Nerinckx, W, Yip, P, Claeyssens, M, Herscovics, A, Howell, P.L. | | Deposit date: | 2007-10-10 | | Release date: | 2008-03-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Modulation of activity by Arg407: structure of a fungal alpha-1,2-mannosidase in complex with a substrate analogue.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2JCO
 
 | | Crystal structure of wild type alpha-1,3 Galactosyltransferase in the absence of ligands | | Descriptor: | GLYCEROL, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYL TRANSFERASE | | Authors: | Jamaluddin, H, Tumbale, P, Withers, S.G, Acharya, K.R, Brew, K. | | Deposit date: | 2006-12-28 | | Release date: | 2007-06-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Conformational Changes Induced by Binding Udp-2F-Galactose to Alpha-1,3 Galactosyltransferase-Implications for Catalysis.
J.Mol.Biol., 369, 2007
|
|
4IID
 
 | | Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus in complex with 1-deoxynojirimycin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1-DEOXYNOJIRIMYCIN, ... | | Authors: | Suzuki, K, Sumitani, J, Kawaguchi, T, Fushinobu, S. | | Deposit date: | 2012-12-20 | | Release date: | 2013-04-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of glycoside hydrolase family 3 beta-glucosidase 1 from Aspergillus aculeatus
Biochem.J., 452, 2013
|
|
