6EVC
 
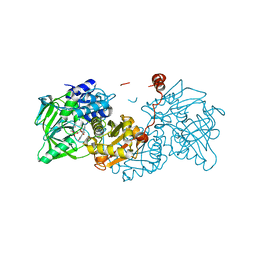 | | Structure of E282Q A. niger Fdc1 in complex with pentafluoro-cinnamic acid | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Ferulic acid decarboxylase 1, MANGANESE (II) ION, ... | | Authors: | Bailey, S.S, David, L, Payne, K.A.P. | | Deposit date: | 2017-11-01 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | The role of conserved residues in Fdc decarboxylase in prenylated flavin mononucleotide oxidative maturation, cofactor isomerization, and catalysis.
J. Biol. Chem., 293, 2018
|
|
2WWY
 
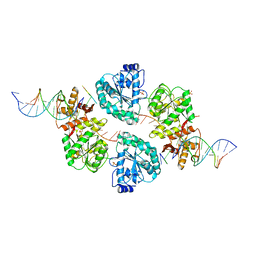 | | Structure of human RECQ-like helicase in complex with a DNA substrate | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*DA DG DC DG DT DC DG DA DG DA DT DC DCP)-3', ATP-DEPENDENT DNA HELICASE Q1, ... | | Authors: | Pike, A.C.W, Zhang, Y, Schnecke, C, Chaikuad, A, Krojer, T, Cooper, C.D.O, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2009-10-30 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Recq1 Helicase-Driven DNA Unwinding, Annealing, and Branch Migration: Insights from DNA Complex Structures
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6C1O
 
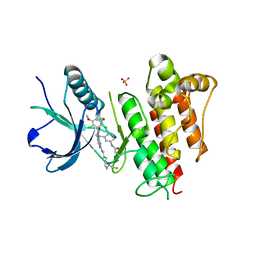 | | FGFR1 kinase domain complexed with FIIN-1 | | Descriptor: | Fibroblast growth factor receptor 1, N-(3-{[3-(2,6-dichloro-3,5-dimethoxyphenyl)-7-{[4-(diethylamino)butyl]amino}-2-oxo-3,4-dihydropyrimido[4,5-d]pyrimidin-1(2H)-yl]methyl}phenyl)prop-2-enamide, SULFATE ION | | Authors: | Kalyukina, M, Yosaatmadja, Y, Smaill, J.B, Squire, C.J. | | Deposit date: | 2018-01-05 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A new class of FGFR1 inhibitors
To Be Published
|
|
1COT
 
 | |
6BHU
 
 | |
4LCT
 
 | | Crystal Structure and Versatile Functional Roles of the COP9 Signalosome Subunit 1 | | Descriptor: | COP9 signalosome complex subunit 1, SULFATE ION | | Authors: | Lee, J.-H, Yi, L, Li, J, Schweitzer, K, Borgmann, M, Naumann, M, Wu, H. | | Deposit date: | 2013-06-23 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and versatile functional roles of the COP9 signalosome subunit 1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5AE9
 
 | | Crystal structure of mouse PI3 kinase delta in complex with GSK2292767 | | Descriptor: | N-[5-[4-(5-{[(2R,6S)-2,6-DIMETHYL-4-MORPHOLINYL]METHYL}-1,3-OXAZOL-2-YL)-1H-INDAZOL-6-YL]-2-(METHYLOXY)-3-PYRIDINYL]METHANESULFONAMIDE, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Down, K.D, Amour, A, Baldwin, I.R, Cooper, A.W.J, Deakin, A.M, Felton, L.M, Guntrip, S.B, Hardy, C, Harrison, Z.A, Jones, K.L, Jones, P, Keeling, S.E, Le, J, Livia, S, Lucas, F, Lunniss, C.J, Parr, N.J, Robinson, E, Rowland, P, Smith, S, Thomas, D.A, Vitulli, G, Washio, Y, Hamblin, N. | | Deposit date: | 2015-08-26 | | Release date: | 2015-09-16 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Optimization of Novel Indazoles as Highly Potent and Selective Inhibitors of Phosphoinositide 3-Kinase Gamma for the Treatment of Respiratory Disease.
J.Med.Chem., 58, 2015
|
|
7D8U
 
 | | Crystal structure of the C-terminal domain of pNP868R from African swine fever virus | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, GTP--RNA guanylyltransferase, ... | | Authors: | Du, X, Geng, Z, Zhang, H. | | Deposit date: | 2020-10-10 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and Biochemical Characteristic of the Methyltransferase (MTase) Domain of RNA Capping Enzyme from African Swine Fever Virus.
J.Virol., 2020
|
|
2V4N
 
 | | Crystal structure of Salmonella typhimurium SurE at 1.7 angstrom resolution in orthorhombic form | | Descriptor: | GLYCEROL, MAGNESIUM ION, MULTIFUNCTIONAL PROTEIN SUR E, ... | | Authors: | Anju, P, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2008-09-26 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Studies on a Mesophilic Stationary Phase Survival Protein (Sur E) from Salmonella Typhimurium
FEBS J., 275, 2008
|
|
1L4F
 
 | | The crystal structure of CobT complexed with 4,5-dimethyl-1,2-phenylenediamine and nicotinate mononucleotide | | Descriptor: | 4,5-DIMETHYL-1,2-PHENYLENEDIAMINE, NICOTINATE MONONUCLEOTIDE, Nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase | | Authors: | Cheong, C.-G, Escalante-Semerena, J, Rayment, I. | | Deposit date: | 2002-03-06 | | Release date: | 2002-09-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Capture of a labile substrate by expulsion of water molecules from the active site of nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT) from Salmonella enterica.
J.Biol.Chem., 277, 2002
|
|
2VBG
 
 | | The complex structure of the branched-chain keto acid decarboxylase (KdcA) from Lactococcus lactis with 2R-1-hydroxyethyl-deazaThDP | | Descriptor: | 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-5-[(1R)-1-HYDROXYETHYL]-3-METHYL-2-THIENYL}ETHYL TRIHYDROGEN DIPHOSPHATE, BRANCHED-CHAIN ALPHA-KETOACID DECARBOXYLASE, MAGNESIUM ION | | Authors: | Berthold, C.L, Gocke, D, Wood, M.D, Leeper, F, Pohl, M, Schneider, G. | | Deposit date: | 2007-09-12 | | Release date: | 2007-11-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Branched-Chain Keto Acid Decarboxylase (Kdca) from Lactococcus Lactis Provides Insights Into the Structural Basis for the Chemo- and Enantioselective Carboligation Reaction
Acta Crystallogr.,Sect.D, 63, 2007
|
|
5SNU
 
 | |
7B6V
 
 | | Sheep Polyomavirus VP1 in complex with 5 mM Forssman antigen pentaose and 20 mM 3'-sialyllactosamine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose, 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Rustmeier, N.H, Stehle, T. | | Deposit date: | 2020-12-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | A novel and highly specific Forssman antigen-binding protein from sheep polyomavirus
Biorxiv, 2023
|
|
1L8P
 
 | | Mg-phosphonoacetohydroxamate complex of S39A yeast enolase 1 | | Descriptor: | MAGNESIUM ION, PHOSPHONOACETOHYDROXAMIC ACID, enolase 1 | | Authors: | Poyner, R.R, Larsen, T.M, Wong, S.W, Reed, G.H. | | Deposit date: | 2002-03-21 | | Release date: | 2002-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and structural changes due to a serine to alanine mutation in the active-site flap of enolase.
Arch.Biochem.Biophys., 401, 2002
|
|
3PXR
 
 | | Apo CDK2 crystallized from Jeffamine | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein kinase 2 | | Authors: | Betzi, S, Alam, R, Schonbrunn, E. | | Deposit date: | 2010-12-10 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a Potential Allosteric Ligand Binding Site in CDK2.
Acs Chem.Biol., 6, 2011
|
|
4AUD
 
 | |
4L8O
 
 | |
5DNC
 
 | |
5E9Z
 
 | | Cytochrome P450 BM3 mutant M11 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Bifunctional cytochrome P450/NADPH--P450 reductase, FE (II) ION, ... | | Authors: | Capoferri, L, Leth, R, ter Haar, E, Mohanty, A.K, Grootenhuis, D.J, Vottero, E, Commandeur, J.N.M, Vermeulen, N.P.E, Jorgensen, F.S, Olsen, L, Geerke, D.P. | | Deposit date: | 2015-10-15 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Insights into regioselective metabolism of mefenamic acid by cytochrome P450 BM3 mutants through crystallography, docking, molecular dynamics, and free energy calculations.
Proteins, 84, 2016
|
|
3IDM
 
 | |
4AU7
 
 | | The structure of the Suv4-20h2 ternary complex with histone H4 | | Descriptor: | 1,2-ETHANEDIOL, HISTONE H4 PEPTIDE, HISTONE-LYSINE N-METHYLTRANSFERASE SUV420H2, ... | | Authors: | Southall, S.M, Cronin, N.B, Wilson, J.R. | | Deposit date: | 2012-05-14 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | A Novel Route to Product Specificity in the Suv4-20 Family of Histone H4K20 Methyltransferases.
Nucleic Acids Res., 42, 2014
|
|
2V8C
 
 | | Mouse Profilin IIa in complex with the proline-rich domain of VASP | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, PROFILIN-2, ... | | Authors: | Kursula, P, Downer, J, Witke, W, Wilmanns, M. | | Deposit date: | 2007-08-06 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | High-Resolution Structural Analysis of Mammalian Profilin 2A Complex Formation with Two Physiological Ligands: The Formin Homology 1 Domain of Mdia1 and the Proline-Rich Domain of Vasp.
J.Mol.Biol., 375, 2008
|
|
4LC8
 
 | | Crystal structure of the mutant H128N of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | Descriptor: | 1,2-ETHANEDIOL, 6-HYDROXYURIDINE-5'-PHOSPHATE, CHLORIDE ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-06-21 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.318 Å) | | Cite: | Crystal structure of the mutant H128N of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP
To be Published
|
|
6BOZ
 
 | | Structure of human SETD8 in complex with covalent inhibitor MS4138 | | Descriptor: | 1,2-ETHANEDIOL, N-(3-{[7-(2-aminoethoxy)-6-methoxy-2-(pyrrolidin-1-yl)quinazolin-4-yl]amino}propyl)prop-2-enamide, N-lysine methyltransferase KMT5A | | Authors: | Babault, N, Anqi, M, Jin, J. | | Deposit date: | 2017-11-21 | | Release date: | 2019-05-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The dynamic conformational landscape of the protein methyltransferase SETD8.
Elife, 8, 2019
|
|
7AE4
 
 | |
