3TDW
 
 | |
3KMH
 
 | |
6M0R
 
 | | 2.7A Yeast Vo state3 | | Descriptor: | (6~{E},10~{E},14~{E},18~{E},22~{E},26~{E},30~{R})-2,6,10,14,18,22,26,30-octamethyldotriaconta-2,6,10,14,18,22,26-heptaene, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, PYROPHOSPHATE, ... | | Authors: | Roh, S.H, Shekhar, M, Pintilie, G, Chipot, C, Wilkens, S, Singharoy, A, Chiu, W. | | Deposit date: | 2020-02-22 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM and MD infer water-mediated proton transport and autoinhibition mechanisms of V o complex.
Sci Adv, 6, 2020
|
|
4CL6
 
 | | Crystal Structure of 3-hydroxydecanoyl-Acyl Carrier Protein Dehydratase (FabA) from Pseudomonas aeruginosa in complex with N-(4- Chlorobenzyl)-3-(2-furyl)-1H-1,2,4-triazol-5-amine | | Descriptor: | 3-HYDROXYDECANOYL-[ACYL-CARRIER-PROTEIN] DEHYDRATASE, N-(4-chlorobenzyl)-5-(furan-2-yl)-1H-1,2,4-triazol-3-amine | | Authors: | Moynie, L, McMahon, S.A, Duthie, F.G, Naismith, J.H. | | Deposit date: | 2014-01-13 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A Substrate Mimic Allows High Throughput Assay of the Faba Protein and Consequently the Identification of a Novel Inhibitor of Pseudomonas Aeruginosa Faba.
J.Mol.Biol., 428, 2016
|
|
6ANV
 
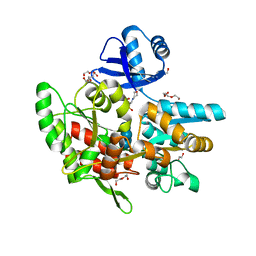 | | Crystal structure of anti-CRISPR protein AcrF1 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, H, Patel, D.J. | | Deposit date: | 2017-08-14 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.265 Å) | | Cite: | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
6Z0K
 
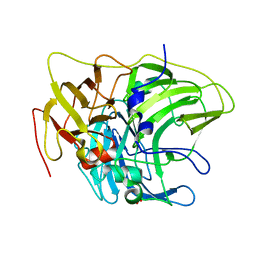 | | Crystal structure of laccase from Pediococcus acidilactici Pp5930 (Hepes pH 7.5) | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, Putative multicopper oxidase mco | | Authors: | Casino, P, Huesa, J, Pardo, I. | | Deposit date: | 2020-05-09 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis and biochemical properties of laccase enzymes from two Pediococcus species.
Microb Biotechnol, 14, 2021
|
|
6Z0J
 
 | |
6ANW
 
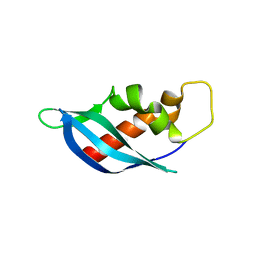 | | Crystal structure of anti-CRISPR protein AcrF10 | | Descriptor: | anti-CRISPR protein AcrF10 | | Authors: | Yang, H, Patel, D.J. | | Deposit date: | 2017-08-14 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.486 Å) | | Cite: | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
6B46
 
 | | Cryo-EM structure of Type I-F CRISPR crRNA-guided Csy surveillance complex with bound anti-CRISPR protein AcrF1 | | Descriptor: | Anti-CRISPR protein AcrF1, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy3, ... | | Authors: | Guo, T.W, Bartesaghi, A, Yang, H, Falconieri, V, Rao, P, Merk, A, Fox, T, Earl, L, Patel, D.J, Subramaniam, S. | | Deposit date: | 2017-09-25 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
6B44
 
 | | Cryo-EM structure of Type I-F CRISPR crRNA-guided Csy surveillance complex with bound target dsDNA | | Descriptor: | CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, CRISPR-associated protein Csy2, ... | | Authors: | Guo, T.W, Bartesaghi, A, Yang, H, Falconieri, V, Rao, P, Merk, A, Fox, T, Earl, L, Patel, D.J, Subramaniam, S. | | Deposit date: | 2017-09-25 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
6B45
 
 | | Cryo-EM structure of Type I-F CRISPR crRNA-guided Csy surveillance complex | | Descriptor: | CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, CRISPR-associated protein Csy2, ... | | Authors: | Guo, T.W, Bartesaghi, A, Yang, H, Falconieri, V, Rao, P, Merk, A, Fox, T, Earl, L, Patel, D.J, Subramaniam, S. | | Deposit date: | 2017-09-25 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
6B48
 
 | | Cryo-EM structure of Type I-F CRISPR crRNA-guided Csy surveillance complex with bound anti-CRISPR protein AcrF10 | | Descriptor: | Anti-CRISPR protein AcrF10, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Guo, T.W, Bartesaghi, A, Yang, H, Falconieri, V, Rao, P, Merk, A, Fox, T, Earl, L, Patel, D.J, Subramaniam, S. | | Deposit date: | 2017-09-25 | | Release date: | 2017-10-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
6B47
 
 | | Cryo-EM structure of Type I-F CRISPR crRNA-guided Csy surveillance complex with bound anti-CRISPR protein AcrF2 | | Descriptor: | Anti-CRISPR protein AcrF2, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Guo, T.W, Bartesaghi, A, Yang, H, Falconieri, V, Rao, P, Merk, A, Fox, T, Earl, L, Patel, D.J, Subramaniam, S. | | Deposit date: | 2017-09-25 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
3ZQ6
 
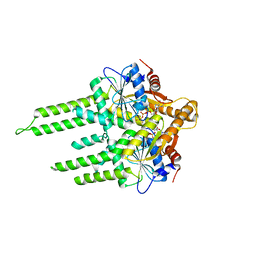 | | ADP-ALF4 COMPLEX OF M. THERM. TRC40 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Sherrill, J, Mariappan, M, Dominik, P, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2011-06-08 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.107 Å) | | Cite: | A Conserved Archaeal Pathway for Tail-Anchored Membrane Protein Insertion.
Traffic, 12, 2011
|
|
5OP0
 
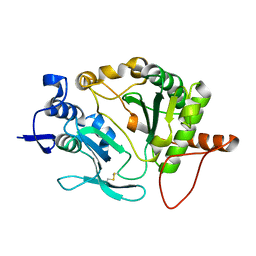 | |
8B7T
 
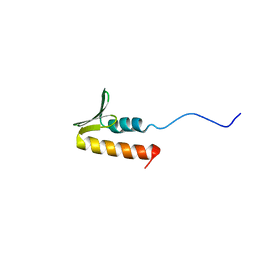 | | CPSF73 CTD3 | | Descriptor: | CPSF73 | | Authors: | Thore, S, Mackereth, C. | | Deposit date: | 2022-10-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-12-06 | | Method: | SOLUTION NMR | | Cite: | Molecular details of the CPSF73-CPSF100 C-terminal heterodimer and interaction with Symplekin.
Open Biology, 13, 2023
|
|
8BA1
 
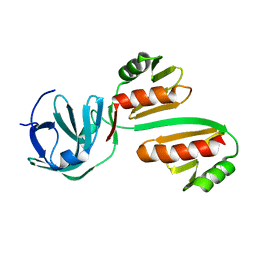 | | CTD12-CTD12 heterodimer from CPSF73 and CPSF100 | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 2, Cleavage and polyadenylation specificity factor subunit 3 | | Authors: | Thore, S, Mackereth, C. | | Deposit date: | 2022-10-10 | | Release date: | 2023-05-03 | | Last modified: | 2023-12-06 | | Method: | SOLUTION NMR | | Cite: | Molecular details of the CPSF73-CPSF100 C-terminal heterodimer and interaction with Symplekin.
Open Biology, 13, 2023
|
|
5IQ9
 
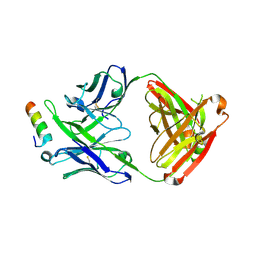 | | Crystal structure of 10E8v4 Fab in complex with an HIV-1 gp41 peptide. | | Descriptor: | 10E8v4 Heavy Chain, 10E8v4 Light Chain, gp41 MPER peptide | | Authors: | Ofek, G, Kwon, Y.D, Caruso, W, Kwong, P.D. | | Deposit date: | 2016-03-10 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Optimization of the Solubility of HIV-1-Neutralizing Antibody 10E8 through Somatic Variation and Structure-Based Design.
J.Virol., 90, 2016
|
|
2FSJ
 
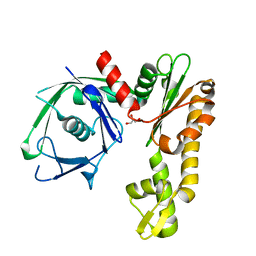 | | Crystal structure of Ta0583, an archaeal actin homolog, native data | | Descriptor: | GLYCEROL, hypothetical protein Ta0583 | | Authors: | Roeben, A, Kofler, C, Nagy, I, Nickell, S, Ulrich Hartl, F, Bracher, A. | | Deposit date: | 2006-01-23 | | Release date: | 2006-04-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an archaeal actin homolog
J.Mol.Biol., 358, 2006
|
|
2FSK
 
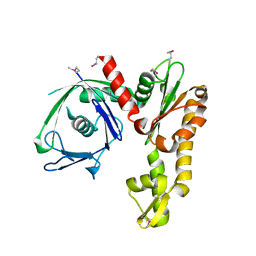 | | Crystal structure of Ta0583, an archaeal actin homolog, SeMet data | | Descriptor: | hypothetical protein Ta0583 | | Authors: | Roeben, A, Kofler, C, Nagy, I, Nickell, S, Ulrich Hartl, F, Bracher, A. | | Deposit date: | 2006-01-23 | | Release date: | 2006-04-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an archaeal actin homolog
J.Mol.Biol., 358, 2006
|
|
8T64
 
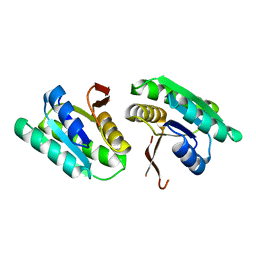 | | Apo Cam1(42-206) | | Descriptor: | Cam1 | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-06-15 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The CRISPR effector Cam1 mediates membrane depolarization for phage defence.
Nature, 625, 2024
|
|
8T66
 
 | | cA6 bound Cam1 | | Descriptor: | Cam1, RNA (5'-R(P*AP*AP*AP*AP*A)-3') | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-06-15 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The CRISPR effector Cam1 mediates membrane depolarization for phage defence.
Nature, 625, 2024
|
|
8T65
 
 | | cA4 bound Cam1 | | Descriptor: | Cam1, RNA (5'-R(P*AP*AP*AP*A)-3') | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-06-15 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The CRISPR effector Cam1 mediates membrane depolarization for phage defence.
Nature, 625, 2024
|
|
2FSN
 
 | | Crystal structure of Ta0583, an archaeal actin homolog, complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, hypothetical protein Ta0583 | | Authors: | Roeben, A, Kofler, C, Nagy, I, Nickell, S, Ulrich Hartl, F, Bracher, A. | | Deposit date: | 2006-01-23 | | Release date: | 2006-04-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of an archaeal actin homolog
J.Mol.Biol., 358, 2006
|
|
5I77
 
 | | Crystal structure of a beta-1,4-endoglucanase from Aspergillus niger | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, Y.J, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-02-17 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Functional and structural analysis of Pichia pastoris-expressed Aspergillus niger 1,4-beta-endoglucanase
Biochem. Biophys. Res. Commun., 475, 2016
|
|
