8SQZ
 
 | |
8SOI
 
 | | Structure of human ULK1 complex core (2:1:1 stoichiometry) | | Descriptor: | Autophagy-related protein 13, RB1-inducible coiled-coil protein 1, Serine/threonine-protein kinase ULK1 | | Authors: | Chen, M, Hurley, J.H. | | Deposit date: | 2023-04-28 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure and activation of the human autophagy-initiating ULK1C:PI3KC3-C1 supercomplex
bioRxiv, 2023
|
|
8SRM
 
 | |
7D0E
 
 | | Crystal structure of FIP200 Claw/p-CCPG1 FIR2 | | Descriptor: | 3-(2-hydroxyethyloxy)-2-[2-(2-hydroxyethyloxy)ethoxymethyl]-2-(2-hydroxyethyloxymethyl)propan-1-ol, Cell cycle progression protein 1 FIR2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhou, Z.X, Pan, L.F. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Phosphorylation regulates the binding of autophagy receptors to FIP200 Claw domain for selective autophagy initiation.
Nat Commun, 12, 2021
|
|
7CZM
 
 | | Crystal structure of FIP200 Claw/p-OPtineurin LIR complex | | Descriptor: | CHLORIDE ION, GLYCEROL, Optineurin LIR, ... | | Authors: | Zhou, Z.X, Pan, L.F. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphorylation regulates the binding of autophagy receptors to FIP200 Claw domain for selective autophagy initiation.
Nat Commun, 12, 2021
|
|
7CZG
 
 | | Crystal structure of FIP200 Claw domain apo form | | Descriptor: | DI(HYDROXYETHYL)ETHER, RB1-inducible coiled-coil protein 1 | | Authors: | Zhou, Z.X, Pan, L.F. | | Deposit date: | 2020-09-08 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Phosphorylation regulates the binding of autophagy receptors to FIP200 Claw domain for selective autophagy initiation.
Nat Commun, 12, 2021
|
|
8YFK
 
 | | Crystal structure of FIP200 claw/TNIP1_FIR_pS123 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MAGNESIUM ION, RB1-inducible coiled-coil protein 1, ... | | Authors: | Lv, M.Q, Wu, S.M. | | Deposit date: | 2024-02-24 | | Release date: | 2024-08-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for TNIP1 binding to FIP200 during mitophagy.
J.Biol.Chem., 300, 2024
|
|
8YFN
 
 | |
8YFL
 
 | | crystal structure of FIP200 claw/TNIP1_FIR_pS122pS123 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, RB1-inducible coiled-coil protein 1, ... | | Authors: | Lv, M.Q, Wu, S.M. | | Deposit date: | 2024-02-24 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for TNIP1 binding to FIP200 during mitophagy.
J.Biol.Chem., 300, 2024
|
|
8YFM
 
 | | Crystal structure of FIP200 claw/TNIP1_FIR_pS122 | | Descriptor: | GLYCEROL, RB1-inducible coiled-coil protein 1, SULFATE ION, ... | | Authors: | Lv, M.Q, Wu, S.M. | | Deposit date: | 2024-02-24 | | Release date: | 2024-08-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for TNIP1 binding to FIP200 during mitophagy.
J.Biol.Chem., 300, 2024
|
|
9C82
 
 | | Structure of human ULK1C:PI3KC3-C1 supercomplex | | Descriptor: | Beclin 1-associated autophagy-related key regulator, Beclin-1, Phosphatidylinositol 3-kinase catalytic subunit type 3, ... | | Authors: | Chen, M, Hurley, J.H. | | Deposit date: | 2024-06-11 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6.84 Å) | | Cite: | Structure and activation of the human autophagy-initiating ULK1C:PI3KC3-C1 supercomplex
bioRxiv, 2023
|
|
2PIQ
 
 | | androgen receptor LBD with small molecule | | Descriptor: | 3-[(4-AMINO-1-TERT-BUTYL-1H-PYRAZOLO[3,4-D]PYRIMIDIN-3-YL)METHYL]PHENOL, 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, ... | | Authors: | Estebanez-Perpina, E, Arnold, A.A, Baxter, J.D, Webb, P, Guy, R.K, Fletterick, K.R. | | Deposit date: | 2007-04-13 | | Release date: | 2007-09-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A surface on the androgen receptor that allosterically regulates coactivator binding.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3L6F
 
 | |
4GBX
 
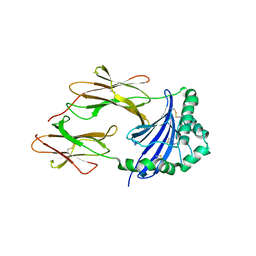 | | Crystal structure of an immune complex at pH 6.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, DM alpha chain, ... | | Authors: | Sethi, D.K, Pos, W, Wucherpfennig, K.W. | | Deposit date: | 2012-07-28 | | Release date: | 2013-01-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of the HLA-DM-HLA-DR1 Complex Defines Mechanisms for Rapid Peptide Selection.
Cell(Cambridge,Mass.), 151, 2012
|
|
4FQX
 
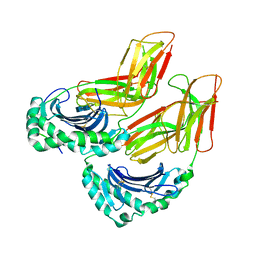 | | Crystal structure of HLA-DM bound to HLA-DR1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, DM alpha chain, ... | | Authors: | Sethi, D.K, Pos, W, Wucherpfennig, K.W. | | Deposit date: | 2012-06-25 | | Release date: | 2013-01-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Crystal Structure of the HLA-DM-HLA-DR1 Complex Defines Mechanisms for Rapid Peptide Selection.
Cell(Cambridge,Mass.), 151, 2012
|
|
4OV5
 
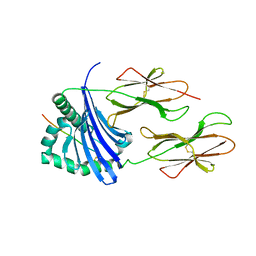 | | Structure of HLA-DR1 with a bound peptide with non-optimal alanine in the P1 pocket | | Descriptor: | HLA class I histocompatibility antigen, A-2 alpha chain, HLA class II histocompatibility antigen, ... | | Authors: | Trenh, P, Yin, L, Stern, L.J. | | Deposit date: | 2014-02-20 | | Release date: | 2014-07-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Susceptibility to HLA-DM Protein Is Determined by a Dynamic Conformation of Major Histocompatibility Complex Class II Molecule Bound with Peptide.
J.Biol.Chem., 289, 2014
|
|
5KNM
 
 | |
6X2W
 
 | | Crystal Structure of PKINES peptide bound to CRM1(E571K) | | Descriptor: | Exportin-1, GLYCEROL, GTP-binding nuclear protein Ran, ... | | Authors: | Baumhardt, J.M. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Recognition of nuclear export signals by CRM1 carrying the oncogenic E571K mutation.
Mol.Biol.Cell, 31, 2020
|
|
1SJE
 
 | | HLA-DR1 complexed with a 16 residue HIV capsid peptide bound in a hairpin conformation | | Descriptor: | Enterotoxin type C-3, GAG polyprotein, HLA class II histocompatibility antigen, ... | | Authors: | Zavala-Ruiz, Z, Strug, I, Walker, B.D, Norris, P.J, Stern, L.J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-08-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A hairpin turn in a class II MHC-bound peptide orients residues outside the binding groove for T cell recognition.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
7O3T
 
 | | I-layer structure (TrwF/VirB9NTD, TrwE/VirB10NTD) of the outer membrane core complex from the fully-assembled R388 type IV secretion system determined by cryo-EM. | | Descriptor: | TrwE protein, TrwF protein | | Authors: | Mace, K, Vadakkepat, A.K, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-04-03 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of a type IV secretion system.
Nature, 607, 2022
|
|
6K60
 
 | | Structural and functional basis for HLA-G isoform recognition of immune checkpoint receptor LILRBs | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, alpha chain G, ... | | Authors: | Kuroki, K, Matsubara, H, Kanda, R, Miyashita, N, Shiroishi, M, Fukunaga, Y, Kamishikiryo, J, Fukunaga, A, Hirose, K, Sugita, Y, Kita, S, Ose, T, Maenaka, K. | | Deposit date: | 2019-05-31 | | Release date: | 2019-11-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.149 Å) | | Cite: | Structural and Functional Basis for LILRB Immune Checkpoint Receptor Recognition of HLA-G Isoforms.
J Immunol., 203, 2019
|
|
3D2U
 
 | | Structure of UL18, a Peptide-Binding Viral MHC Mimic, Bound to a Host Inhibitory Receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Actin, Beta-2-microglobulin, ... | | Authors: | Yang, Z, Bjorkman, P.J. | | Deposit date: | 2008-05-08 | | Release date: | 2008-07-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of UL18, a peptide-binding viral MHC mimic, bound to a host inhibitory receptor
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4OJK
 
 | | Structure of the cGMP Dependent Protein Kinase II and Rab11b Complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Rab-11B, cGMP-dependent protein kinase 2 | | Authors: | Reger, A.S, Yang, M.P, Guo, E, Kim, C. | | Deposit date: | 2014-01-21 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.657 Å) | | Cite: | Crystal Structure of the cGMP-dependent Protein Kinase II Leucine Zipper and Rab11b Protein Complex Reveals Molecular Details of G-kinase-specific Interactions.
J.Biol.Chem., 289, 2014
|
|
7O3J
 
 | | O-layer structure (TrwH/VirB7, TrwF/VirB9CTD, TrwE/VirB10CTD) of the outer membrane core complex from the fully-assembled R388 type IV secretion system determined by cryo-EM. | | Descriptor: | TrwE protein, TrwF protein, TrwH protein | | Authors: | Mace, K, Vadakkepat, A.K, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-04-01 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Cryo-EM structure of a type IV secretion system.
Nature, 607, 2022
|
|
3S4S
 
 | | Crystal structure of CD4 mutant bound to HLA-DR1 | | Descriptor: | GLYCEROL, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Li, Y. | | Deposit date: | 2011-05-20 | | Release date: | 2011-09-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Affinity maturation of human CD4 by yeast surface display and crystal structure of a CD4-HLA-DR1 complex.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
