5KZH
 
 | |
5L2F
 
 | | High Resolution Structure of Acinetobacter baumannii beta-lactamase OXA-51 I129L/K83D bound to doripenem | | Descriptor: | (4R,5S)-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-3-({(3S,5S)-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl}sulfanyl)-4,5-dihydro-1H-pyrrole-2-carboxylic acid, ACETATE ION, Beta-lactamase, ... | | Authors: | June, C.M, Powers, R.A, Leonard, D.A, Muckenthaler, T. | | Deposit date: | 2016-08-01 | | Release date: | 2016-09-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The structure of a doripenem-bound OXA-51 class D beta-lactamase variant with enhanced carbapenemase activity.
Protein Sci., 25, 2016
|
|
5LP5
 
 | | Complex between Penicillin-Binding Protein (PBP2) and MreC from Helicobacter pylori | | Descriptor: | Penicillin-binding protein 2 (Pbp2), Rod shape-determining protein (MreC) | | Authors: | Contreras-Martel, C, Martins, A, Ecobichon, C, Maragno, D.M, Mattei, P.J, El Ghachi, M, Hicham, S, Hardouin, P, Boneca, I.G, Dessen, A. | | Deposit date: | 2016-08-11 | | Release date: | 2017-08-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Molecular architecture of the PBP2-MreC core bacterial cell wall synthesis complex.
Nat Commun, 8, 2017
|
|
5LP4
 
 | | Penicillin-Binding Protein (PBP2) from Helicobacter pylori | | Descriptor: | Penicillin-binding protein 2 (Pbp2), SULFATE ION | | Authors: | Contreras-Martel, C, Martins, A, Ecobichon, C, Maragno, D.M, Mattei, P.J, El Ghachi, M, Boneca, I.G, Dessen, A. | | Deposit date: | 2016-08-11 | | Release date: | 2017-08-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Molecular architecture of the PBP2-MreC core bacterial cell wall synthesis complex.
Nat Commun, 8, 2017
|
|
5TG4
 
 | | OXA-24/40 in Complex with Boronic Acid BA16 | | Descriptor: | BICARBONATE ION, Beta-lactamase, D-Glyceraldehyde, ... | | Authors: | Powers, R.A, Werner, J.P, Mitchell, J.M. | | Deposit date: | 2016-09-27 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Exploring the potential of boronic acids as inhibitors of OXA-24/40 beta-lactamase.
Protein Sci., 26, 2017
|
|
5TG6
 
 | | OXA-24/40 in Complex with Boronic Acid BA4 | | Descriptor: | BICARBONATE ION, Beta-lactamase, SULFATE ION, ... | | Authors: | Powers, R.A, Werner, J.P, Mitchell, J.M. | | Deposit date: | 2016-09-27 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Exploring the potential of boronic acids as inhibitors of OXA-24/40 beta-lactamase.
Protein Sci., 26, 2017
|
|
5TG5
 
 | | OXA-24/40 in Complex with Boronic Acid BA8 | | Descriptor: | BICARBONATE ION, Beta-lactamase, METHANETHIOL, ... | | Authors: | Powers, R.A, Werner, J.P, Mitchell, J.M. | | Deposit date: | 2016-09-27 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Exploring the potential of boronic acids as inhibitors of OXA-24/40 beta-lactamase.
Protein Sci., 26, 2017
|
|
5TG7
 
 | | OXA-24/40 in Complex with Boronic Acid BA3 | | Descriptor: | (3-{[(furan-2-yl)methyl]carbamoyl}phenyl)boronic acid, BICARBONATE ION, Beta-lactamase, ... | | Authors: | Powers, R.A, Werner, J.P, Mitchell, J.M. | | Deposit date: | 2016-09-27 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Exploring the potential of boronic acids as inhibitors of OXA-24/40 beta-lactamase.
Protein Sci., 26, 2017
|
|
5M18
 
 | | Crystal structure of PBP2a from MRSA in the presence of Cefepime ligand | | Descriptor: | CADMIUM ION, Penicillin-binding protein 2, beta-muramic acid | | Authors: | Molina, R, Batuecas, M.T, Hermoso, J.A. | | Deposit date: | 2016-10-07 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Conformational Dynamics in Penicillin-Binding Protein 2a of Methicillin-Resistant Staphylococcus aureus, Allosteric Communication Network and Enablement of Catalysis.
J. Am. Chem. Soc., 139, 2017
|
|
5M1A
 
 | | Crystal structure of PBP2a from MRSA in the presence of Ceftazidime ligand | | Descriptor: | CADMIUM ION, Penicillin-binding protein 2, beta-muramic acid | | Authors: | Molina, R, Batuecas, M.T, Hermoso, J.A. | | Deposit date: | 2016-10-07 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Dynamics in Penicillin-Binding Protein 2a of Methicillin-Resistant Staphylococcus aureus, Allosteric Communication Network and Enablement of Catalysis.
J. Am. Chem. Soc., 139, 2017
|
|
5M19
 
 | | Crystal structure of PBP2a from MRSA in the presence of Oxacillin ligand | | Descriptor: | CADMIUM ION, Penicillin-binding protein 2, beta-muramic acid | | Authors: | Molina, R, Batuecas, M.T, Hermoso, J.A. | | Deposit date: | 2016-10-07 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Dynamics in Penicillin-Binding Protein 2a of Methicillin-Resistant Staphylococcus aureus, Allosteric Communication Network and Enablement of Catalysis.
J. Am. Chem. Soc., 139, 2017
|
|
5TRO
 
 | | 1.8 Angstrom Resolution Crystal Structure of Dimerization and Transpeptidase domains (residues 39-608) of Penicillin-Binding Protein 1 from Staphylococcus aureus. | | Descriptor: | CHLORIDE ION, Penicillin-binding protein 1 | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-26 | | Release date: | 2016-11-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Resolution Crystal Structure of Dimerization and Transpeptidase domains (residues 39-608) of Penicillin-Binding Protein 1 from Staphylococcus aureus.
To Be Published
|
|
5U2G
 
 | | 2.6 Angstrom Resolution Crystal Structure of Penicillin-Binding Protein 1A from Haemophilus influenzae | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-30 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | 2.6 Angstrom Resolution Crystal Structure of Penicillin-Binding Protein 1A from Haemophilus influenzae.
To Be Published
|
|
5U47
 
 | | 1.95 Angstrom Resolution Crystal Structure of Penicillin Binding Protein 2X from Streptococcus thermophilus | | Descriptor: | ACETATE ION, CHLORIDE ION, Penicillin binding protein 2X | | Authors: | Minasov, G, Shuvalova, L, Cardona-Correa, A, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-03 | | Release date: | 2016-12-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Resolution Crystal Structure of Penicillin Binding Protein 2X from Streptococcus thermophilus.
To Be Published
|
|
5MMY
 
 | | Crystal structure of OXA10 with HEPES | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase OXA-10, ... | | Authors: | Brem, J. | | Deposit date: | 2016-12-12 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | (13)C-Carbamylation as a mechanistic probe for the inhibition of class D beta-lactamases by avibactam and halide ions.
Org. Biomol. Chem., 15, 2017
|
|
5MNU
 
 | | OXA-10 Avibactam complex with bound bromide | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, BROMIDE ION, Beta-lactamase OXA-10, ... | | Authors: | Brem, J, McDonough, M, Clifton, I. | | Deposit date: | 2016-12-13 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | (13)C-Carbamylation as a mechanistic probe for the inhibition of class D beta-lactamases by avibactam and halide ions.
Org. Biomol. Chem., 15, 2017
|
|
5MOX
 
 | | OXA-10 Avibactam complex with bound CO2 | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase OXA-10, CARBON DIOXIDE, ... | | Authors: | Brem, J. | | Deposit date: | 2016-12-14 | | Release date: | 2017-07-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | (13)C-Carbamylation as a mechanistic probe for the inhibition of class D beta-lactamases by avibactam and halide ions.
Org. Biomol. Chem., 15, 2017
|
|
5MOZ
 
 | | OXA-10 Avibactam complex with bound Iodide | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase OXA-10, CHLORIDE ION, ... | | Authors: | Brem, J. | | Deposit date: | 2016-12-15 | | Release date: | 2017-07-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | (13)C-Carbamylation as a mechanistic probe for the inhibition of class D beta-lactamases by avibactam and halide ions.
Org. Biomol. Chem., 15, 2017
|
|
5UY7
 
 | |
5VFD
 
 | | Diazabicyclooctenone ETX2514 bound to Class D beta lactamase OXA-24 from A. baumannii | | Descriptor: | (2S,5R)-1-formyl-4-methyl-5-[(sulfooxy)amino]-1,2,5,6-tetrahydropyridine-2-carboxamide, (2S,5R)-4-methyl-7-oxo-6-(sulfooxy)-1,6-diazabicyclo[3.2.1]oct-3-ene-2-carboxamide, Beta-lactamase, ... | | Authors: | Olivier, N.B, Lahiri, S. | | Deposit date: | 2017-04-07 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | ETX2514 is a broad-spectrum beta-lactamase inhibitor for the treatment of drug-resistant Gram-negative bacteria including Acinetobacter baumannii.
Nat Microbiol, 2, 2017
|
|
5OAU
 
 | |
5ODZ
 
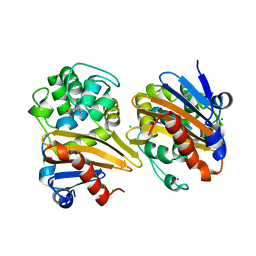 | | CRYSTAL STRUCTURE OF THE BETA-LACTAMASE OXA-163 | | Descriptor: | Beta-lactamase, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Lund, B.A, Carlsen, T.J.O, Leiros, H.K.S. | | Deposit date: | 2017-07-07 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure, activity and thermostability investigations of OXA-163, OXA-181 and OXA-245 using biochemical analysis, crystal structures and differential scanning calorimetry analysis.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5OE2
 
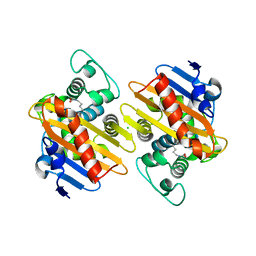 | | CRYSTAL STRUCTURE OF THE BETA-LACTAMASE OXA-245 | | Descriptor: | Beta-lactamase, CHLORIDE ION, SODIUM ION | | Authors: | Lund, B.A, Leiros, H.K.S. | | Deposit date: | 2017-07-07 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure, activity and thermostability investigations of OXA-163, OXA-181 and OXA-245 using biochemical analysis, crystal structures and differential scanning calorimetry analysis.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5OE0
 
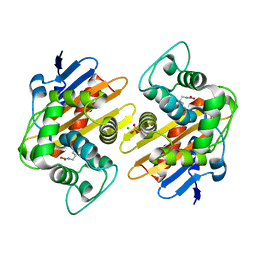 | | CRYSTAL STRUCTURE OF THE BETA-LACTAMASE OXA-181 | | Descriptor: | Beta-lactamase, CHLORIDE ION, SULFATE ION | | Authors: | Lund, B.A, Carlsen, T.J.O, Leiros, H.K.S, Thomassen, A.M. | | Deposit date: | 2017-07-07 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.0500083 Å) | | Cite: | Structure, activity and thermostability investigations of OXA-163, OXA-181 and OXA-245 using biochemical analysis, crystal structures and differential scanning calorimetry analysis.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
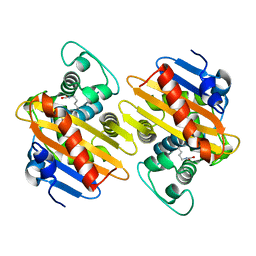
5OFT
 
 | |
