2HK3
 
 | |
2M56
 
 | | The structure of the complex of cytochrome P450cam and its electron donor putidaredoxin determined by paramagnetic NMR spectroscopy | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Hiruma, Y, Hass, M.A.S, Ubbink, M. | | Deposit date: | 2013-02-14 | | Release date: | 2013-08-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of the cytochrome p450cam-putidaredoxin complex determined by paramagnetic NMR spectroscopy and crystallography.
J.Mol.Biol., 425, 2013
|
|
2LYG
 
 | | Fuc_TBA | | Descriptor: | 2-hydroxyethyl 6-deoxy-beta-L-galactopyranoside, DNA (5'-D(P*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3') | | Authors: | Gomez-Pinto, I, Vengut-Climent, E, Lucas, R, Avio, A, Eritja, R, Gonzalez-Ibaez, C, Morales, J. | | Deposit date: | 2012-09-18 | | Release date: | 2014-01-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Carbohydrate-DNA interactions at G-quadruplexes: folding and stability changes by attaching sugars at the 5'-end.
Chemistry, 19, 2013
|
|
6S7N
 
 | |
5AXH
 
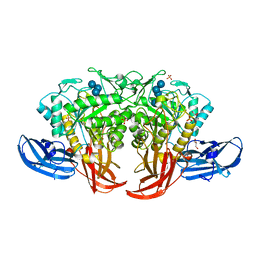 | | Crystal structure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus, D312G mutant in complex with isomaltohexaose | | Descriptor: | Dextranase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Suzuki, N, Kishine, N, Fujimoto, Z, Sakurai, M, Momma, M, Ko, J.A, Nam, S.H, Kimura, A, Kim, Y.M. | | Deposit date: | 2015-07-29 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus
J.Biochem., 159, 2016
|
|
2RHS
 
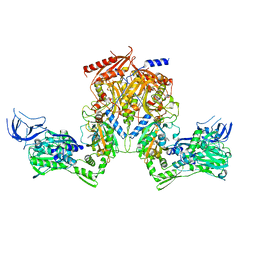 | | PheRS from Staphylococcus haemolyticus- rational protein engineering and inhibitor studies | | Descriptor: | 1-{3-[(4-pyridin-2-ylpiperazin-1-yl)sulfonyl]phenyl}-3-(1,3-thiazol-2-yl)urea, Phenylalanyl-tRNA synthetase alpha chain, Phenylalanyl-tRNA synthetase beta chain, ... | | Authors: | Evdokimov, A.G, Mekel, M. | | Deposit date: | 2007-10-09 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational protein engineering in action: the first crystal structure of a phenylalanine tRNA synthetase from Staphylococcus haemolyticus.
J.Struct.Biol., 162, 2008
|
|
6S0P
 
 | |
7AOB
 
 | | Crystal structure of Thermaerobacter marianensis malate dehydrogenase | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Bertrand, Q, Lasalle, L, Girard, E, Madern, D. | | Deposit date: | 2020-10-14 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of Thermaerobacter marianensis malate dehydrogenase
To Be Published
|
|
2MEE
 
 | |
2LY4
 
 | | HMGB1-facilitated p53 DNA binding occurs via HMG-box/p53 transactivation domain interaction and is regulated by the acidic tail | | Descriptor: | Cellular tumor antigen p53, High mobility group protein B1 | | Authors: | Rowell, J.P, Simpson, K.L, Stott, K, Watson, M, Thomas, J.O. | | Deposit date: | 2012-09-12 | | Release date: | 2012-10-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | HMGB1-Facilitated p53 DNA Binding Occurs via HMG-Box/p53 Transactivation Domain Interaction, Regulated by the Acidic Tail.
Structure, 20, 2012
|
|
2LZ2
 
 | |
2M41
 
 | |
5AOV
 
 | |
5APD
 
 | | Hen Egg White Lysozyme not illuminated with 0.4THz radiation | | Descriptor: | LYSOZYME C, SODIUM ION | | Authors: | Lundholm, I, Rodilla, H, Wahlgren, W.Y, Duelli, A, Bourenkov, G, Vukusic, J, Friedman, R, Stake, J, Schneider, T, Katona, G. | | Deposit date: | 2015-09-15 | | Release date: | 2016-01-13 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Terahertz Radiation Induces Non-Thermal Structural Changes Associated with Frohlich Condensation in a Protein Crystal
Struct.Dyn., 2, 2015
|
|
2LXH
 
 | | NMR structure of the RING domain in ubiquitin ligase gp78 | | Descriptor: | E3 ubiquitin-protein ligase AMFR, ZINC ION | | Authors: | Das, R, Linag, Y, Mariano, J, Li, J, Huang, T, King, A, Weissman, A, Ji, X, Byrd, R. | | Deposit date: | 2012-08-27 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine.
Embo J., 32, 2013
|
|
5B06
 
 | | Lysozyme (denatured by NaOD and refolded) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Kita, A, Morimoto, Y. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Effective Deuterium Exchange Method for Neutron Crystal Structure Analysis with Unfolding-Refolding Processes
Mol Biotechnol., 58, 2016
|
|
7AX7
 
 | |
5B1G
 
 | |
5B2E
 
 | | N,N'-diacetylchitobiose deacetylase from Pyrococcus horikoshii complexed with its inhibitor MPG (acetate-containing condition) | | Descriptor: | 2-deoxy-2-{[(S)-hydroxy(methyl)phosphoryl]amino}-beta-D-glucopyranose, HEXANE-1,6-DIOL, Putative uncharacterized protein PH0499, ... | | Authors: | Nakamura, T, Niiyama, M, Ida, K, Uegaki, K. | | Deposit date: | 2016-01-15 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate recognition of N,N'-diacetylchitobiose deacetylase from Pyrococcus horikoshii
J.Struct.Biol., 195, 2016
|
|
7AEY
 
 | | Salmonella typhimurium neuraminidase in complex with isocarba-DANA. | | Descriptor: | (3~{S},4~{S},5~{R})-4-acetamido-3-oxidanyl-5-[(1~{S},2~{R})-1,2,3-tris(oxidanyl)propyl]cyclohexane-1-carboxylic acid, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Salinger, M.T, Kuhn, P, Laver, W.G, Pape, T, Schneider, T.R, Sheldrick, G.M, Vasella, A.T, Vimr, E.R, Vorwerk, S, Garman, E.F. | | Deposit date: | 2020-09-18 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.919 Å) | | Cite: | Salmonella typhimurium neuraminidase in complex with isocarba-DANA.
To Be Published
|
|
6S6T
 
 | |
5Y9V
 
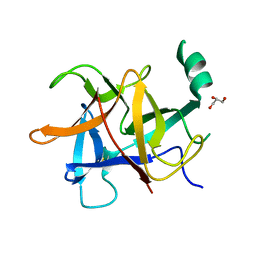 | | Crystal structure of diamondback moth ryanodine receptor N-terminal domain | | Descriptor: | CHLORIDE ION, GLYCEROL, Ryanodine receptor 1 | | Authors: | Lin, L, Yuchi, Z. | | Deposit date: | 2017-08-28 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.841 Å) | | Cite: | Crystal structure of ryanodine receptor N-terminal domain from Plutella xylostella reveals two potential species-specific insecticide-targeting sites.
Insect Biochem. Mol. Biol., 92, 2017
|
|
7AG8
 
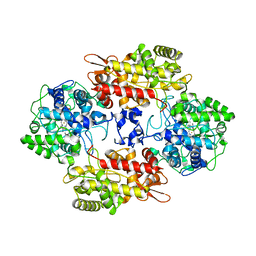 | |
2BOA
 
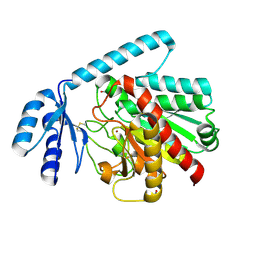 | | Human procarboxypeptidase A4. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBOXYPEPTIDASE A4, ... | | Authors: | Garcia-Castellanos, R, Bonet-Figueredo, R, Pallares, I, Ventura, S, Aviles, F.X, Vendrell, J, Gomis-Ruth, F.X. | | Deposit date: | 2005-04-08 | | Release date: | 2005-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Detailed Molecular Comparison between the Inhibition Mode of A/B-Type Carboxypeptidases in the Zymogen State and by the Endogenous Inhibitor Latexin.
Cell.Mol.Life Sci., 62, 2005
|
|
5B59
 
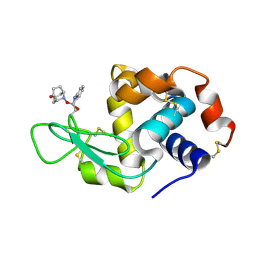 | | Hen egg-white lysozyme modified with a keto-ABNO. | | Descriptor: | (2~{S})-2-azanyl-3-[(2~{R},3~{S})-2-oxidanyl-3-[[(1~{S},5~{R})-3-oxidanylidene-9-azabicyclo[3.3.1]nonan-9-yl]oxy]-1,2-dihydroindol-3-yl]propanal, Lysozyme C | | Authors: | Sasaki, D, Seki, Y, Sohma, Y, Oisaki, K, Kanai, M. | | Deposit date: | 2016-04-28 | | Release date: | 2016-09-14 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Transition Metal-Free Tryptophan-Selective Bioconjugation of Proteins
J.Am.Chem.Soc., 138, 2016
|
|
