2AH7
 
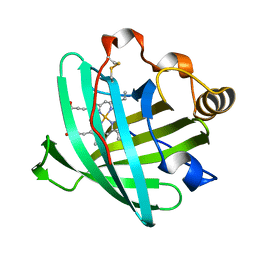 | | Crystal structure of nitrophorin 2 aqua complex | | Descriptor: | Nitrophorin 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Weichsel, A, Berry, R.E, Walker, F.A, Montfort, W.R. | | Deposit date: | 2005-07-27 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures, ligand induced conformational change and heme deformation in complexes of nitrophorin 2, a nitric oxide transport protein from rhodnius prolixus
To be Published
|
|
2AMM
 
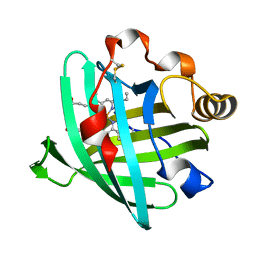 | | Crystal structure of L122V/L132V mutant of nitrophorin 2 | | Descriptor: | Nitrophorin 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Weichsel, A, Berry, R.E, Walker, F.A, Montfort, W.R. | | Deposit date: | 2005-08-09 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures, ligand induced conformational change and heme deformation in complexes of nitrophorin 2, a nitric oxide transport protein from rhodnius prolixus
To be Published
|
|
1Q5L
 
 | | NMR structure of the substrate binding domain of DnaK bound to the peptide NRLLLTG | | Descriptor: | Chaperone protein dnaK, peptide NRLLLTG | | Authors: | Stevens, S.Y, Cai, S, Pellecchia, M, Zuiderweg, E.R. | | Deposit date: | 2003-08-08 | | Release date: | 2003-11-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the bacterial HSP70 chaperone protein domain DnaK(393-507) in complex with the peptide NRLLLTG.
Protein Sci., 12, 2003
|
|
1PPE
 
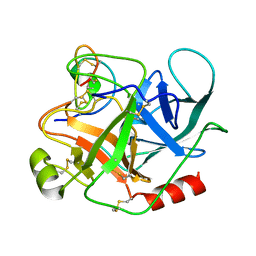 | |
1PT2
 
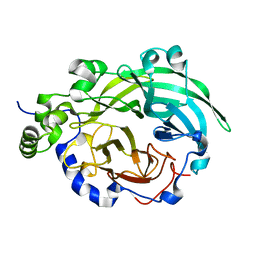 | |
2AG5
 
 | | Crystal Structure of Human DHRS6 | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, dehydrogenase/reductase (SDR family) member 6 | | Authors: | Kunde, G, Lukacik, P, Papagrigoriou, E, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A, Von Delft, F, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-07-26 | | Release date: | 2005-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Characterization of human DHRS6, an orphan short chain dehydrogenase/reductase enzyme: a novel, cytosolic type 2 R-beta-hydroxybutyrate dehydrogenase
J.Biol.Chem., 281, 2006
|
|
1X8N
 
 | | 1.08 A Crystal Structure Of Nitrophorin 4 From Rhodnius Prolixus Complexed With Nitric Oxide at pH 7.4 | | Descriptor: | NITRIC OXIDE, Nitrophorin 4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kondrashov, D.A, Roberts, S.A, Weichsel, A, Montfort, W.R. | | Deposit date: | 2004-08-18 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Protein functional cycle viewed at atomic resolution: conformational change and mobility in nitrophorin 4 as a function of pH and NO binding
Biochemistry, 43, 2004
|
|
2ALL
 
 | | Crystal structure of L122V/L132V mutant of nitrophorin 2 | | Descriptor: | Nitrophorin 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Weichsel, A, Berry, R.E, Walker, F.A, Montfort, W.R. | | Deposit date: | 2005-08-05 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal structures, ligand induced conformational change and heme deformation in complexes of nitrophorin 2, a nitric oxide transport protein from rhodnius prolixus
To be Published
|
|
1RPJ
 
 | | CRYSTAL STRUCTURE OF D-ALLOSE BINDING PROTEIN FROM ESCHERICHIA COLI | | Descriptor: | PROTEIN (PRECURSOR OF PERIPLASMIC SUGAR RECEPTOR), SULFATE ION, ZINC ION, ... | | Authors: | Chaudhuri, B, Jones, T.A, Mowbray, S.L. | | Deposit date: | 1999-02-04 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of D-allose binding protein from Escherichia coli bound to D-allose at 1.8 A resolution.
J.Mol.Biol., 286, 1999
|
|
1ILX
 
 | | Excited State Dynamics in Photosystem II Revised. New Insights from the X-ray Structure. | | Descriptor: | 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, CADMIUM ION, CHLOROPHYLL A, ... | | Authors: | Vasilev, S, Orth, P, Zouni, A, Owens, T.G, Bruce, D. | | Deposit date: | 2001-05-09 | | Release date: | 2001-07-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Excited-state dynamics in photosystem II: insights from the x-ray crystal structure.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1RIE
 
 | | STRUCTURE OF A WATER SOLUBLE FRAGMENT OF THE RIESKE IRON-SULFUR PROTEIN OF THE BOVINE HEART MITOCHONDRIAL CYTOCHROME BC1-COMPLEX | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, RIESKE IRON-SULFUR PROTEIN | | Authors: | Iwata, S, Saynovits, M, Link, T.A, Michel, H. | | Deposit date: | 1996-02-23 | | Release date: | 1996-12-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a water soluble fragment of the 'Rieske' iron-sulfur protein of the bovine heart mitochondrial cytochrome bc1 complex determined by MAD phasing at 1.5 A resolution.
Structure, 4, 1996
|
|
1SIG
 
 | |
1JD6
 
 | | Crystal Structure of DIAP1-BIR2/Hid Complex | | Descriptor: | APOPTOSIS 1 INHIBITOR, ZINC ION, head involution defective protein | | Authors: | Wu, J.W, Cocina, A.E, Chai, J, Hay, B.A, Shi, Y. | | Deposit date: | 2001-06-12 | | Release date: | 2001-12-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of a functional DIAP1 fragment bound to grim and hid peptides.
Mol.Cell, 8, 2001
|
|
1JD4
 
 | | Crystal Structure of DIAP1-BIR2 | | Descriptor: | APOPTOSIS 1 INHIBITOR, ZINC ION | | Authors: | Wu, J.W, Cocina, A.E, Chai, J, Hay, B.A, Shi, Y. | | Deposit date: | 2001-06-12 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of a functional DIAP1 fragment bound to grim and hid peptides.
Mol.Cell, 8, 2001
|
|
2ATI
 
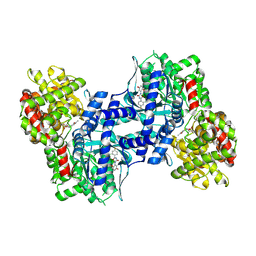 | | Glycogen Phosphorylase Inhibitors | | Descriptor: | Glycogen phosphorylase, liver form, N-(2-CHLORO-4-FLUOROBENZOYL)-N'-(5-HYDROXY-2-METHOXYPHENYL)UREA, ... | | Authors: | Klabunde, T, Wendt, K.U, Kadereit, D, Brachvogel, V, Burger, H.J, Herling, A.W, Oikonomakos, N.G, Schmoll, D, Sarubbi, E, von Roedern, E, Schoenafinger, K, Defossa, E. | | Deposit date: | 2005-08-25 | | Release date: | 2006-08-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Acyl ureas as human liver glycogen phosphorylase inhibitors for the treatment of type 2 diabetes.
J.Med.Chem., 48, 2005
|
|
1NIF
 
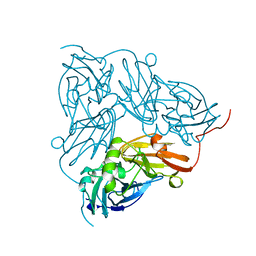 | | THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Adman, E.T, Godden, J.W, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted.
J.Biol.Chem., 270, 1995
|
|
1NIB
 
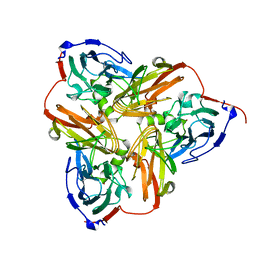 | | THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Adman, E.T, Godden, J.W, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted.
J.Biol.Chem., 270, 1995
|
|
1NKQ
 
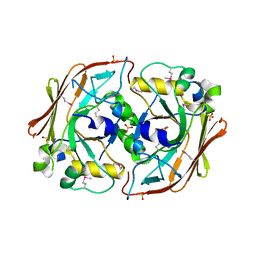 | | Crystal structure of yeast ynq8, a fumarylacetoacetate hydrolase family protein | | Descriptor: | ACETIC ACID, CALCIUM ION, Hypothetical 28.8 kDa protein in PSD1-SKO1 intergenic region, ... | | Authors: | Eswaramoorthy, S, Kumaran, D, Daniels, B, Studier, F.W, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-01-03 | | Release date: | 2004-06-15 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crtystal Structure of Yeast Hypothetical Protein YNQ8_YEAST
To be Published
|
|
1NIE
 
 | | THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Adman, E.T, Godden, J.W, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted.
J.Biol.Chem., 270, 1995
|
|
1YWD
 
 | | 1.08 A Structure of Ferrous NP4 (aquo complex) | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, nitrophorin 4 | | Authors: | Maes, E.M, Weichsel, A, Roberts, S.A, Montfort, W.R. | | Deposit date: | 2005-02-17 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Ultrahigh Resolution Structures of Nitrophorin 4: Heme Distortion in Ferrous CO and NO Complexes
Biochemistry, 44, 2005
|
|
2BLZ
 
 | | RNAse after a high dose X-ray "burn" | | Descriptor: | CHLORIDE ION, RIBONUCLEASE PANCREATIC | | Authors: | Nanao, M.H, Ravelli, R.B. | | Deposit date: | 2005-03-08 | | Release date: | 2005-09-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Improving Radiation-Damage Substructures for Rip.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1E7R
 
 | | GDP 4-keto-6-deoxy-D-mannose epimerase reductase Y136E | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETYLPHOSPHATE, GDP-FUCOSE SYNTHETASE, ... | | Authors: | Rosano, C, Izzo, G, Bolognesi, M. | | Deposit date: | 2000-09-07 | | Release date: | 2000-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing the Catalytic Mechanism of Gdp-4-Keto-6-Deoxy-D-Mannose Epimerase/Reductase by Kinetic and Crystallographic Characterization of Site-Specific Mutants
J.Mol.Biol., 303, 2000
|
|
1ELY
 
 | | E. COLI ALKALINE PHOSPHATASE MUTANT (S102C) | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Stec, B, Hehir, M, Brennan, C, Nolte, M, Kantrowitz, E.R. | | Deposit date: | 1998-02-10 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Kinetic and X-ray structural studies of three mutant E. coli alkaline phosphatases: insights into the catalytic mechanism without the nucleophile Ser102.
J.Mol.Biol., 277, 1998
|
|
1EXY
 
 | | SOLUTION STRUCTURE OF HTLV-1 PEPTIDE BOUND TO ITS RNA APTAMER TARGET | | Descriptor: | HTLV-1 REX PEPTIDE, RNA APTAMER, 33-MER | | Authors: | Jiang, F, Gorin, A, Hu, W, Majumdar, A, Baskerville, S, Xu, W, Ellington, A, Patel, D.J. | | Deposit date: | 2000-05-05 | | Release date: | 2000-05-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Anchoring an extended HTLV-1 Rex peptide within an RNA major groove containing junctional base triples.
Structure Fold.Des., 7, 1999
|
|
1ELX
 
 | | E. COLI ALKALINE PHOSPHATASE MUTANT (S102A) | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Stec, B, Hehir, M, Brennan, C, Nolte, M, Kantrowitz, E.R. | | Deposit date: | 1998-02-10 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Kinetic and X-ray structural studies of three mutant E. coli alkaline phosphatases: insights into the catalytic mechanism without the nucleophile Ser102.
J.Mol.Biol., 277, 1998
|
|
