1BWL
 
 | | OLD YELLOW ENZYME (OYE1) DOUBLE MUTANT H191N:N194H | | Descriptor: | FLAVIN MONONUCLEOTIDE, PROTEIN (NADPH DEHYDROGENASE 1) | | Authors: | Brown, B.J, Deng, Z, Karplus, P.A, Massey, V. | | Deposit date: | 1998-09-24 | | Release date: | 1998-09-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | On the active site of Old Yellow Enzyme. Role of histidine 191 and asparagine 194.
J.Biol.Chem., 273, 1998
|
|
1BX1
 
 | | FERREDOXIN:NADP+ OXIDOREDUCTASE (FERREDOXIN REDUCTASE) MUTANT E312Q | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, PROTEIN (FERREDOXIN:NADP+ OXIDOREDUCTASE), ... | | Authors: | Aliverti, A, Deng, Z, Ravasi, D, Piubelli, L, Karplus, P.A, Zanetti, G. | | Deposit date: | 1998-10-10 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the function of the invariant glutamyl residue 312 in spinach ferredoxin-NADP+ reductase.
J.Biol.Chem., 273, 1998
|
|
1BY4
 
 | | STRUCTURE AND MECHANISM OF THE HOMODIMERIC ASSEMBLY OF THE RXR ON DNA | | Descriptor: | DNA (5'-D(*C*TP*AP*GP*GP*TP*CP*AP*AP*AP*GP*GP*TP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*AP*CP*CP*TP*TP*TP*GP*AP*CP*CP*TP*A)-3'), PROTEIN (RETINOIC ACID RECEPTOR RXR-ALPHA), ... | | Authors: | Zhao, Q, Chasse, S.A, Devarakonda, S, Sierk, M.L, Ahvazi, B, Sigler, P.B, Rastinejad, F. | | Deposit date: | 1998-10-22 | | Release date: | 2000-01-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of RXR-DNA interactions.
J.Mol.Biol., 296, 2000
|
|
1BYB
 
 | | CRYSTAL STRUCTURES OF SOYBEAN BETA-AMYLASE REACTED WITH BETA-MALTOSE AND MALTAL: ACTIVE SITE COMPONENTS AND THEIR APPARENT ROLE IN CATALYSIS | | Descriptor: | BETA-AMYLASE, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Mikami, B, Degano, M, Hehre, E.J, Sacchettini, J.C. | | Deposit date: | 1994-01-25 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of soybean beta-amylase reacted with beta-maltose and maltal: active site components and their apparent roles in catalysis.
Biochemistry, 33, 1994
|
|
1BYW
 
 | |
1BZX
 
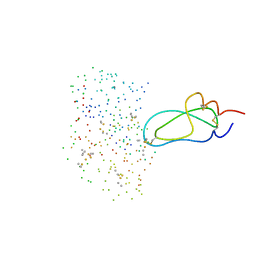 | | THE CRYSTAL STRUCTURE OF ANIONIC SALMON TRYPSIN IN COMPLEX WITH BOVINE PANCREATIC TRYPSIN INHIBITOR | | Descriptor: | CALCIUM ION, PROTEIN (BOVINE PANCREATIC TRYPSIN INHIBITOR), PROTEIN (TRYPSIN) | | Authors: | Helland, R, Leiros, I, Berglund, G.I, Willassen, N.P, Smalas, A.O. | | Deposit date: | 1998-11-05 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of anionic salmon trypsin in complex with bovine pancreatic trypsin inhibitor.
Eur.J.Biochem., 256, 1998
|
|
1C0E
 
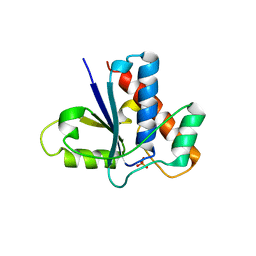 | | Active Site S19A Mutant of Bovine Heart Phosphotyrosyl Phosphatase | | Descriptor: | PHOSPHATE ION, PROTEIN (TYROSINE PHOSPHATASE (ORTHOPHOSPHORIC MONOESTER PHOSPHOHYDROLASE)) | | Authors: | Tabernero, L, Evans, B.N, Tishmack, P.A, Van Etten, R.L, Stauffacher, C.V. | | Deposit date: | 1999-07-15 | | Release date: | 1999-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of the bovine protein tyrosine phosphatase dimer reveals a potential self-regulation mechanism.
Biochemistry, 38, 1999
|
|
1C1C
 
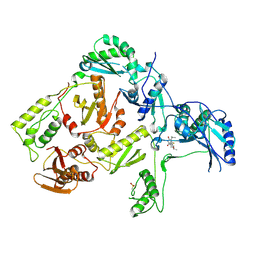 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH TNK-6123 | | Descriptor: | 6-(cyclohexylsulfanyl)-1-(ethoxymethyl)-5-(1-methylethyl)pyrimidine-2,4(1H,3H)-dione, HIV-1 REVERSE TRANSCRIPTASE (A-CHAIN), HIV-1 REVERSE TRANSCRIPTASE (B-CHAIN) | | Authors: | Hopkins, A.L, Ren, J, Tanaka, H, Baba, M, Okamato, M, Stuart, D.I, Stammers, D.K. | | Deposit date: | 1999-07-21 | | Release date: | 2000-07-21 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design of MKC-442 (emivirine) analogues with improved activity against drug-resistant HIV mutants.
J.Med.Chem., 42, 1999
|
|
1C2D
 
 | |
1BJI
 
 | |
1BK1
 
 | | ENDO-1,4-BETA-XYLANASE C | | Descriptor: | ENDO-1,4-B-XYLANASE C | | Authors: | Fushinobu, S, Ito, K, Konno, M, Wakagi, T, Matsuzawa, H. | | Deposit date: | 1998-07-14 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and mutational analyses of an extremely acidophilic and acid-stable xylanase: biased distribution of acidic residues and importance of Asp37 for catalysis at low pH.
Protein Eng., 11, 1998
|
|
1BKD
 
 | | COMPLEX OF HUMAN H-RAS WITH HUMAN SOS-1 | | Descriptor: | H-RAS, SON OF SEVENLESS-1 | | Authors: | Boriack-Sjodin, P.A, Margarit, S.M, Sagi, D.B, Kuriyan, J. | | Deposit date: | 1998-07-06 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis of the activation of Ras by Sos.
Nature, 394, 1998
|
|
1BL4
 
 | | FKBP MUTANT F36V COMPLEXED WITH REMODELED SYNTHETIC LIGAND | | Descriptor: | PROTEIN (FK506 BINDING PROTEIN), {3-[3-(3,4-DIMETHOXY-PHENYL)-1-(1-{1-[2-(3,4,5-TRIMETHOXY-PHENYL)-BUTYRYL]-PIPERIDIN-2YL}-VINYLOXY)-PROPYL]-PHENOXY}-ACETIC ACID | | Authors: | Hatada, M.H, Clackson, T, Yang, W, Rozamus, L.W, Amara, J, Rollins, C.T, Stevenson, L.F, Magari, S.R, Wood, S.A, Courage, N.L, Lu, X, Cerasoli Junior, F, Gilman, M, Holt, D. | | Deposit date: | 1998-07-23 | | Release date: | 1998-09-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redesigning an FKBP-ligand interface to generate chemical dimerizers with novel specificity.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1B0K
 
 | | S642A:FLUOROCITRATE COMPLEX OF ACONITASE | | Descriptor: | CITRATE ANION, IRON/SULFUR CLUSTER, OXYGEN ATOM, ... | | Authors: | Lloyd, S.J, Lauble, H, Prasad, G.S, Stout, C.D. | | Deposit date: | 1998-11-11 | | Release date: | 1999-11-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The mechanism of aconitase: 1.8 A resolution crystal structure of the S642a:citrate complex.
Protein Sci., 8, 1999
|
|
1B18
 
 | |
1B1I
 
 | | CRYSTAL STRUCTURE OF HUMAN ANGIOGENIN | | Descriptor: | CITRIC ACID, HYDROLASE ANGIOGENIN | | Authors: | Leonidas, D.D, Acharya, K.R. | | Deposit date: | 1998-11-20 | | Release date: | 1999-04-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined crystal structures of native human angiogenin and two active site variants: implications for the unique functional properties of an enzyme involved in neovascularisation during tumour growth.
J.Mol.Biol., 285, 1999
|
|
1AU0
 
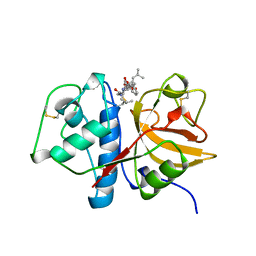 | | CRYSTAL STRUCTURE OF THE CYSTEINE PROTEASE HUMAN CATHEPSIN K IN COMPLEX WITH A COVALENT SYMMETRIC DIACYLAMINOMETHYL KETONE INHIBITOR | | Descriptor: | 1,3-BIS[[N-[(PHENYLMETHOXY)CARBONYL]-L-LEUCYL]AMINO]-2-PROPANONE, CATHEPSIN K | | Authors: | Zhao, B, Smith, W.W, Janson, C.A, Abdel-Meguid, S.S. | | Deposit date: | 1997-09-09 | | Release date: | 1998-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Design of Potent and Selective Cathepsin K Inhibitors
J.Am.Chem.Soc., 119, 1997
|
|
1AU8
 
 | | HUMAN CATHEPSIN G | | Descriptor: | CATHEPSIN G, N-(3-carboxypropanoyl)-L-valyl-N-[(1R)-5-amino-1-phosphonopentyl]-L-prolinamide | | Authors: | Medrano, F.J, Bode, W, Banbula, A, Potempa, J. | | Deposit date: | 1997-09-12 | | Release date: | 1998-10-14 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | HUMAN CATHEPSIN G
to be published
|
|
1AON
 
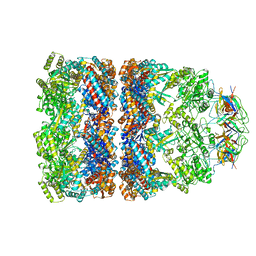 | | CRYSTAL STRUCTURE OF THE ASYMMETRIC CHAPERONIN COMPLEX GROEL/GROES/(ADP)7 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GROEL, GROEL/GROES COMPLEX, ... | | Authors: | Xu, Z, Horwich, A.L, Sigler, P.B. | | Deposit date: | 1997-07-08 | | Release date: | 1997-10-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex.
Nature, 388, 1997
|
|
1AVL
 
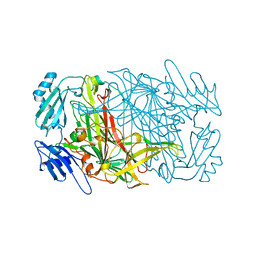 | |
1B24
 
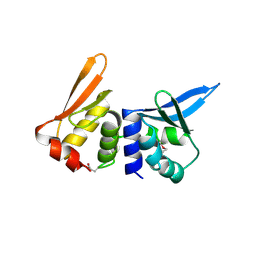 | | I-DMOI, INTRON-ENCODED ENDONUCLEASE | | Descriptor: | Homing endonuclease I-DmoI | | Authors: | Van Roey, P, Silva, G.H. | | Deposit date: | 1998-12-03 | | Release date: | 1999-03-24 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the thermostable archaeal intron-encoded endonuclease I-DmoI.
J.Mol.Biol., 286, 1999
|
|
1BAI
 
 | | Crystal structure of Rous sarcoma virus protease in complex with inhibitor | | Descriptor: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE | | Authors: | Wu, J, Adomat, J.M, Ridky, T.W, Louis, J.M, Leis, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 1998-04-17 | | Release date: | 1999-01-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for specificity of retroviral proteases.
Biochemistry, 37, 1998
|
|
1AW9
 
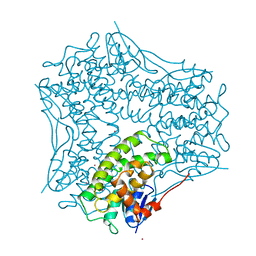 | | STRUCTURE OF GLUTATHIONE S-TRANSFERASE III IN APO FORM | | Descriptor: | CADMIUM ION, GLUTATHIONE S-TRANSFERASE III | | Authors: | Neuefeind, T, Huber, R, Reinemer, P, Knaeblein, J. | | Deposit date: | 1997-10-13 | | Release date: | 1998-10-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cloning, sequencing, crystallization and X-ray structure of glutathione S-transferase-III from Zea mays var. mutin: a leading enzyme in detoxification of maize herbicides.
J.Mol.Biol., 274, 1997
|
|
1BAW
 
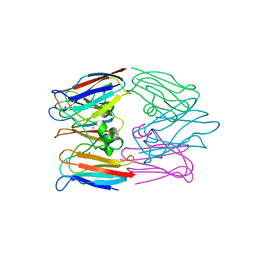 | | PLASTOCYANIN FROM PHORMIDIUM LAMINOSUM | | Descriptor: | COPPER (II) ION, PLASTOCYANIN, ZINC ION | | Authors: | Bond, C.S, Guss, J.M, Freeman, H.C, Wagner, M.J, Howe, C.J, Bendall, D.S. | | Deposit date: | 1998-04-19 | | Release date: | 1999-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of plastocyanin from the cyanobacterium Phormidium laminosum.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1BD7
 
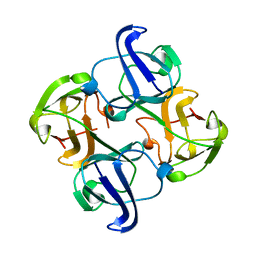 | | CIRCULARLY PERMUTED BB2-CRYSTALLIN | | Descriptor: | CIRCULARLY PERMUTED BB2-CRYSTALLIN | | Authors: | Wright, G, Basak, A.K, Mayr, E.-M, Glockshuber, R, Slingsby, C. | | Deposit date: | 1998-05-12 | | Release date: | 1998-10-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Circular permutation of betaB2-crystallin changes the hierarchy of domain assembly.
Protein Sci., 7, 1998
|
|
