3T81
 
 | |
6FAS
 
 | | Crystal structure of VAL1 B3 domain in complex with cognate DNA | | Descriptor: | B3 domain-containing transcription repressor VAL1, DNA (5'-D(*AP*GP*CP*CP*AP*TP*GP*CP*AP*CP*CP*G)-3'), DNA (5'-D(*CP*GP*GP*TP*GP*CP*AP*TP*GP*GP*CP*T)-3') | | Authors: | Sasnauskas, G. | | Deposit date: | 2017-12-17 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of DNA target recognition by the B3 domain of Arabidopsis epigenome reader VAL1.
Nucleic Acids Res., 46, 2018
|
|
3TG0
 
 | | E. coli alkaline phosphatase with bound inorganic phosphate | | Descriptor: | Alkaline phosphatase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Bobyr, E, Lassila, J.K, Wiersma-Koch, H.I, Fenn, T.D, Lee, J.J, Nikolic-Hughes, I, Hodgson, K.O, Rees, D.C, Hedman, B, Herschlag, D. | | Deposit date: | 2011-08-16 | | Release date: | 2011-11-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution analysis of Zn(2+) coordination in the alkaline phosphatase superfamily by EXAFS and x-ray crystallography.
J.Mol.Biol., 415, 2012
|
|
3THH
 
 | |
4G0J
 
 | |
3THY
 
 | | Human MutSbeta complexed with an IDL of 2 bases (Loop2) and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA Loop2 minus strand, DNA Loop2 plus strand, ... | | Authors: | Yang, W. | | Deposit date: | 2011-08-19 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Mechanism of mismatch repair revealed by human MutSbeta bound to unpaired DNA loops
Nat.Struct.Mol.Biol., 19, 2012
|
|
3THW
 
 | | Human MutSbeta complexed with an IDL of 4 bases (Loop4) and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA Loop4 hairpin, DNA mismatch repair protein Msh2, ... | | Authors: | Yang, W. | | Deposit date: | 2011-08-19 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Mechanism of mismatch recognition revealed by human MutSbeta bound to unpaired DNA loops
Nat.Struct.Mol.Biol., 19, 2012
|
|
4G5X
 
 | | Crystal structures of N-acyl homoserine lactonase AidH | | Descriptor: | Alpha/beta hydrolase fold protein | | Authors: | Liang, D.C, Yan, X.X, Gao, A. | | Deposit date: | 2012-07-18 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | High-resolution structures of AidH complexes provide insights into a novel catalytic mechanism for N-acyl homoserine lactonase
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6EXC
 
 | |
3S8R
 
 | | Crystal Structures of Glutaryl 7-Aminocephalosporanic Acid Acylase: Insight into Autoproteolytic Activation | | Descriptor: | GLYCEROL, Glutaryl-7-aminocephalosporanic-acid acylase | | Authors: | Kim, J.K, Yang, I.S, Park, S.S, Kim, K.H. | | Deposit date: | 2011-05-30 | | Release date: | 2011-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of glutaryl 7-aminocephalosporanic acid acylase: insight into autoproteolytic activation.
Biochemistry, 42, 2003
|
|
1VC2
 
 | |
3SEP
 
 | | E. coli (lacZ) beta-galactosidase (S796A) | | Descriptor: | Beta-galactosidase, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | Deposit date: | 2011-06-10 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Ser-796 of Beta-Galactosidase (E. coli) Plays a Key Role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
4G09
 
 | |
5CL4
 
 | |
4G8C
 
 | |
1ULV
 
 | | Crystal Structure of Glucodextranase Complexed with Acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, CALCIUM ION, glucodextranase | | Authors: | Mizuno, M, Tonozuka, T, Suzuki, S, Uotsu-Tomita, R, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2003-09-16 | | Release date: | 2003-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural insights into substrate specificity and function of glucodextranase
J.Biol.Chem., 279, 2004
|
|
1UMC
 
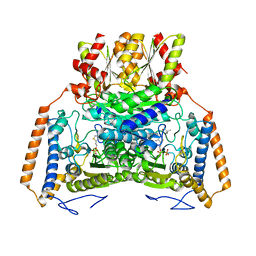 | | branched-chain 2-oxo acid dehydrogenase (E1) from Thermus thermophilus HB8 with 4-methylpentanoate | | Descriptor: | 2-oxo acid dehydrogenase alpha subunit, 2-oxo acid dehydrogenase beta subunit, 4-METHYL VALERIC ACID, ... | | Authors: | Nakai, T, Nakagawa, N, Maoka, N, Masui, R, Kuramitsu, S, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-25 | | Release date: | 2004-03-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ligand-induced Conformational Changes and a Reaction Intermediate in Branched-chain 2-Oxo Acid Dehydrogenase (E1) from Thermus thermophilus HB8, as Revealed by X-ray Crystallography
J.Mol.Biol., 337, 2004
|
|
1UN1
 
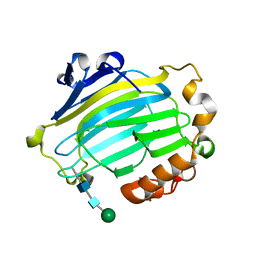 | | Xyloglucan endotransglycosylase native structure. | | Descriptor: | GOLD ION, XYLOGLUCAN ENDOTRANSGLYCOSYLASE, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Johansson, P, Brumer, H, Kallas, A, Henriksson, H, Denman, S, Teeri, T.T, Jones, T.A. | | Deposit date: | 2003-09-03 | | Release date: | 2004-03-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of a Poplar Xyloglucan Endotransglycosylase Reveal Details of Transglycosylation Acceptor Binding
Plant Cell, 16, 2004
|
|
3M6D
 
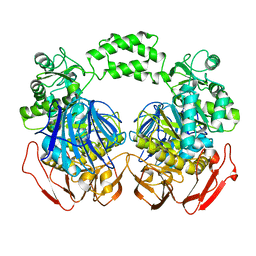 | | The crystal structure of the d307a mutant of glycoside Hydrolase (family 31) from ruminococcus obeum atcc 29174 | | Descriptor: | Uncharacterized protein | | Authors: | Tan, K, Tesar, C, Freeman, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-15 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Novel alpha-glucosidase from human gut microbiome: substrate specificities and their switch.
Faseb J., 24, 2010
|
|
5CM3
 
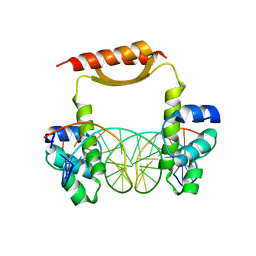 | | Crystal Structure of KorA, a plasmid-encoded, global transcription regulator | | Descriptor: | 5'-D(CP*CP*AP*AP*GP*TP*TP*TP*AP*GP*CP*TP*AP*AP*AP*CP*TP*TP*GP*GP*)-3', TrfB transcriptional repressor protein | | Authors: | White, S.A, Hyde, E.I, Rajasekar, K.V. | | Deposit date: | 2015-07-16 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Flexibility of KorA, a plasmid-encoded, global transcription regulator, in the presence and the absence of its operator.
Nucleic Acids Res., 44, 2016
|
|
1UXK
 
 | | Large improvement in the thermal stability of a tetrameric malate dehydrogenase by single point mutations at the dimer-dimer interface | | Descriptor: | CADMIUM ION, CHLORIDE ION, MALATE DEHYDROGENASE, ... | | Authors: | Bjork, A, Dalhus, B, Mantzilas, D, Eijsink, V.G.H, Sirevag, R. | | Deposit date: | 2004-02-25 | | Release date: | 2004-08-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Large Improvement in the Thermal Stability of a Tetrameric Malate Dehydrogenase by Single Point Mutations at the Dimer-Dimer Interface.
J.Mol.Biol., 341, 2004
|
|
3SCW
 
 | | Crystal Structure of Rice BGlu1 E386G/Y341A Mutant Complexed with Cellotetraose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, SULFATE ION, ... | | Authors: | Pengthaisong, S, Withers, S.G, Kuaprasert, B, Ketudat Cairns, J.R. | | Deposit date: | 2011-06-08 | | Release date: | 2012-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural investigation of the basis for cellooligosaccharide synthesis by rice BGlu1 glycosynthases
to be published
|
|
1UQY
 
 | | Xylanase Xyn10B mutant (E262S) from Cellvibrio mixtus in complex with xylopentaose | | Descriptor: | ENDOXYLANASE, MAGNESIUM ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose, ... | | Authors: | Pell, G, Taylor, E.J, Gloster, T.M, Turkenburg, J.P, Fontes, C.M.G.A, Ferreira, L.M.A, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2003-10-23 | | Release date: | 2003-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The Mechanisms by which Family 10 Glycoside Hydrolases Bind Decorated Substrates
J.Biol.Chem., 279, 2004
|
|
4GZT
 
 | | N2 neuraminidase D151G mutant of A/Tanzania/205/2010 H3N2 in complex with oseltamivir carboxylate | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2012-09-06 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Influenza virus neuraminidases with reduced enzymatic activity that avidly bind sialic Acid receptors.
J.Virol., 86, 2012
|
|
3ON3
 
 | | The crystal structure of keto/oxoacid ferredoxin oxidoreductase, gamma subunit from Geobacter sulfurreducens PCA | | Descriptor: | Keto/oxoacid ferredoxin oxidoreductase, gamma subunit, SULFATE ION | | Authors: | Tan, K, Zhang, R, Hatzos, C, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-27 | | Release date: | 2010-09-22 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | The crystal structure of keto/oxoacid ferredoxin oxidoreductase, gamma subunit from Geobacter sulfurreducens PCA
To be Published
|
|
