1G6K
 
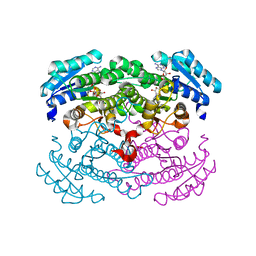 | | Crystal structure of glucose dehydrogenase mutant E96A complexed with NAD+ | | Descriptor: | GLUCOSE 1-DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamamoto, K, Kurisu, G, Kusunoki, M, Tabata, S, Urabe, I, Osaki, S. | | Deposit date: | 2000-11-06 | | Release date: | 2003-08-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of stability-increasing mutants of glucose dehydrogenase
To be Published
|
|
3UKY
 
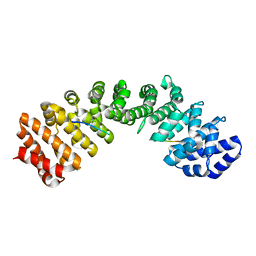 | | Mouse importin alpha: yeast CBP80 cNLS complex | | Descriptor: | Importin subunit alpha-2, Nuclear cap-binding protein complex subunit 1 | | Authors: | Marfori, M, Forwood, J.K, Lonhienne, T.G, Kobe, B. | | Deposit date: | 2011-11-10 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of High-Affinity Nuclear Localization Signal Interactions with Importin-alpha
Traffic, 13, 2012
|
|
1AOU
 
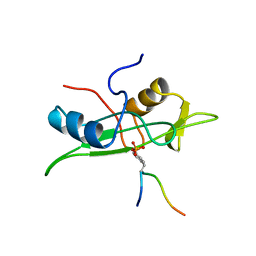 | | NMR STRUCTURE OF THE FYN SH2 DOMAIN COMPLEXED WITH A PHOSPHOTYROSYL PEPTIDE, 22 STRUCTURES | | Descriptor: | FYN PROTEIN-TYROSINE KINASE, PHOSPHOTYROSYL PEPTIDE | | Authors: | Mulhern, T.D, Shaw, G.L, Morton, C.J, Day, A.J, Campbell, I.D. | | Deposit date: | 1997-07-10 | | Release date: | 1998-01-14 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | The SH2 domain from the tyrosine kinase Fyn in complex with a phosphotyrosyl peptide reveals insights into domain stability and binding specificity.
Structure, 5, 1997
|
|
1ATB
 
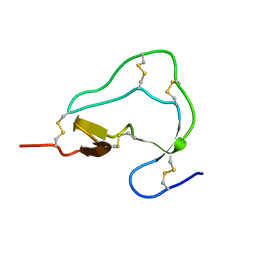 | |
1ATD
 
 | |
2B3Y
 
 | | Structure of a monoclinic crystal form of human cytosolic aconitase (IRP1) | | Descriptor: | ACETATE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Dupuy, J, Fontecilla-Camps, J.C, Volbeda, A. | | Deposit date: | 2005-09-22 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of human iron regulatory protein 1 as cytosolic aconitase
Structure, 14, 2006
|
|
2AME
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 N14Q | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-18 | | Release date: | 1999-04-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
1BDC
 
 | | STAPHYLOCOCCUS AUREUS PROTEIN A, IMMUNOGLOBULIN-BINDING B DOMAIN, NMR, 10 STRUCTURES | | Descriptor: | STAPHYLOCOCCUS AUREUS PROTEIN A | | Authors: | Gouda, H, Torigoe, H, Saito, A, Sato, M, Arata, Y, Shimada, I. | | Deposit date: | 1996-06-28 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the B domain of staphylococcal protein A: comparisons of the solution and crystal structures.
Biochemistry, 31, 1992
|
|
2AHN
 
 | | High resolution structure of a cherry allergen Pru av 2 | | Descriptor: | Thaumatin-like protein | | Authors: | Dall'Antonia, Y, Pavkov, T, Fuchs, H, Breiteneder, H, Keller, W. | | Deposit date: | 2005-07-28 | | Release date: | 2006-10-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The high-resolution crystal structure of an allergenic thaumatin-like protein, Pru av 2, isolated from ripe cherries
To be published
|
|
1BGQ
 
 | |
1BD0
 
 | | ALANINE RACEMASE COMPLEXED WITH ALANINE PHOSPHONATE | | Descriptor: | ALANINE RACEMASE, {1-[(3-HYDROXY-METHYL-5-PHOSPHONOOXY-METHYL-PYRIDIN-4-YLMETHYL)-AMINO]-ETHYL}-PHOSPHONIC ACID | | Authors: | Stamper, G.F, Morollo, A.A, Ringe, D. | | Deposit date: | 1998-05-12 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Reaction of alanine racemase with 1-aminoethylphosphonic acid forms a stable external aldimine.
Biochemistry, 37, 1998
|
|
1IDK
 
 | | PECTIN LYASE A | | Descriptor: | PECTIN LYASE A | | Authors: | Mayans, O, Scott, M, Connerton, I, Gravesen, T, Benen, J, Visser, J, Pickersgill, R, Jenkins, J. | | Deposit date: | 1996-10-04 | | Release date: | 1997-10-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Two crystal structures of pectin lyase A from Aspergillus reveal a pH driven conformational change and striking divergence in the substrate-binding clefts of pectin and pectate lyases.
Structure, 5, 1997
|
|
3TDT
 
 | |
2LGN
 
 | | Lactococcin 972 | | Descriptor: | Lactococcin 972 | | Authors: | Turner, D.L, Lamosa, P, Martinez, B. | | Deposit date: | 2011-07-29 | | Release date: | 2012-08-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and properties of lactococcin 972 from Lactococcus lactis
To be Published
|
|
1AYR
 
 | | ARRESTIN FROM BOVINE ROD OUTER SEGMENTS | | Descriptor: | ARRESTIN | | Authors: | Granzin, J, Wilden, U, Choe, H.-W, Labahn, J, Krafft, B, Bueldt, G. | | Deposit date: | 1997-11-10 | | Release date: | 1998-11-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | X-ray crystal structure of arrestin from bovine rod outer segments.
Nature, 391, 1998
|
|
1ILZ
 
 | | OUTER MEMBRANE PHOSPHOLIPASE A FROM ESCHERICHIA COLI N156A ACTIVE SITE MUTANT pH 6.1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, OUTER MEMBRANE PHOSPHOLIPASE A, octyl beta-D-glucopyranoside | | Authors: | Snijder, H.J, Van Eerde, J.H, Kingma, R.L, Kalk, K.H, Dekker, N, Egmond, M.R, Dijkstra, B.W. | | Deposit date: | 2001-05-09 | | Release date: | 2001-10-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural investigations of the active-site mutant Asn156Ala of outer membrane phospholipase A: function of the Asn-His interaction in the catalytic triad.
Protein Sci., 10, 2001
|
|
1DN8
 
 | |
1ITI
 
 | | THE HIGH RESOLUTION THREE-DIMENSIONAL SOLUTION STRUCTURE OF HUMAN INTERLEUKIN-4 DETERMINED BY MULTI-DIMENSIONAL HETERONUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | INTERLEUKIN-4 | | Authors: | Clore, G.M, Powers, B, Garrett, D.S, Gronenborn, A.M. | | Deposit date: | 1993-04-12 | | Release date: | 1993-07-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The high-resolution, three-dimensional solution structure of human interleukin-4 determined by multidimensional heteronuclear magnetic resonance spectroscopy.
Biochemistry, 32, 1993
|
|
2APJ
 
 | | X-Ray Structure of Protein from Arabidopsis Thaliana AT4G34215 at 1.6 Angstrom Resolution | | Descriptor: | Putative Esterase | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Mccoy, J.G, Bitto, E, Bingman, C.A, Allard, S.T, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-08-16 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure at 1.6 Angstroms resolution of the protein product of the At4g34215 gene from Arabidopsis thaliana.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1DVE
 
 | | CRYSTAL STRUCTURE OF RAT HEME OXYGENASE-1 IN COMPLEX WITH HEME | | Descriptor: | HEME OXYGENASE-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Omata, Y, Kakuta, Y, Sakamoto, H, Noguchi, M, Fukuyama, K. | | Deposit date: | 2000-01-20 | | Release date: | 2000-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of rat heme oxygenase-1 in complex with heme.
FEBS Lett., 471, 2000
|
|
1DWR
 
 | | MYOGLOBIN (HORSE HEART) WILD-TYPE COMPLEXED WITH CO | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Chu, K, Vojtechovsky, J, McMahon, B.H, Sweet, R.M, Berendzen, J, Schlichting, I. | | Deposit date: | 1999-12-11 | | Release date: | 2000-03-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of a New Ligand Binding Intermediate in Wildtype Carbonmonoxy Myoglobin
Nature, 403, 2000
|
|
1IIU
 
 | |
1IE7
 
 | | PHOSPHATE INHIBITED BACILLUS PASTEURII UREASE CRYSTAL STRUCTURE | | Descriptor: | NICKEL (II) ION, PHOSPHATE ION, UREASE ALPHA SUBUNIT, ... | | Authors: | Benini, S, Rypniewski, W.R, Wilson, K.S, Ciurli, S, Mangani, S. | | Deposit date: | 2001-04-09 | | Release date: | 2001-04-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-based rationalization of urease inhibition by phosphate: novel insights into the enzyme mechanism.
J.Biol.Inorg.Chem., 6, 2001
|
|
1E2B
 
 | | NMR STRUCTURE OF THE C10S MUTANT OF ENZYME IIB CELLOBIOSE OF THE PHOSPHOENOL-PYRUVATE DEPENDENT PHOSPHOTRANSFERASE SYSTEM OF ESCHERICHIA COLI, 17 STRUCTURES | | Descriptor: | ENZYME IIB-CELLOBIOSE | | Authors: | Ab, E, Schuurman-Wolters, G, Reizer, J, Saier, M.H, Dijkstra, K, Scheek, R.M, Robillard, G.T. | | Deposit date: | 1996-11-15 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR side-chain assignments and solution structure of enzyme IIBcellobiose of the phosphoenolpyruvate-dependent phosphotransferase system of Escherichia coli.
Protein Sci., 6, 1997
|
|
1CEA
 
 | |
