4M59
 
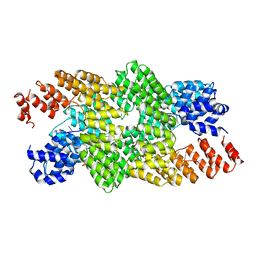 | | Crystal structure of the pentatricopeptide repeat protein PPR10 in complex with an 18-nt psaJ RNA element | | Descriptor: | Chloroplast pentatricopeptide repeat protein 10, PHOSPHATE ION, psaJ RNA | | Authors: | Yin, P, Li, Q, Yan, C, Liu, Y, Yan, N. | | Deposit date: | 2013-08-08 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural basis for the modular recognition of single-stranded RNA by PPR proteins.
Nature, 504, 2013
|
|
4HIU
 
 | |
5F7Z
 
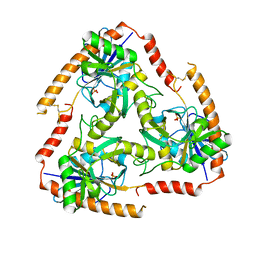 | | Crystal structure of Double Mutant S12T and N87T of Adenosine/Methylthioadenosine Phosphorylase from Schistosoma mansoni in APO Form | | Descriptor: | Methylthioadenosine phosphorylase, PHOSPHATE ION | | Authors: | Torini, J.R, Brandao-Neto, J, DeMarco, R, Pereira, H.M. | | Deposit date: | 2015-12-08 | | Release date: | 2016-12-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Schistosoma mansoni Adenosine Phosphorylase/5'-Methylthioadenosine Phosphorylase and Its Importance on Adenosine Salvage Pathway.
PLoS Negl Trop Dis, 10, 2016
|
|
3FP9
 
 | |
2ZYR
 
 | | A. Fulgidus lipase with fatty acid fragment and magnesium | | Descriptor: | Lipase, putative, MAGNESIUM ION, ... | | Authors: | Chen, C.K, Ko, T.P, Guo, R.T, Wang, A.H. | | Deposit date: | 2009-01-28 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure of the alkalohyperthermophilic Archaeoglobus fulgidus lipase contains a unique C-terminal domain essential for long-chain substrate binding.
J.Mol.Biol., 390, 2009
|
|
2DPR
 
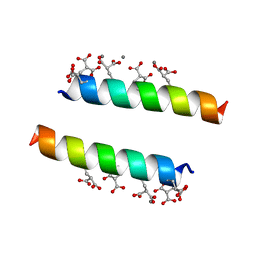 | | The crystal structures of the calcium-bound con-G and con-T(K7Glu) dimeric peptides demonstrate a novel metal-dependent helix-forming motif | | Descriptor: | CALCIUM ION, Conantokin-T | | Authors: | Cnudde, S.E, Prorok, M, Dai, Q, Castellino, F.J, Geiger, J.H. | | Deposit date: | 2006-05-13 | | Release date: | 2007-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structures of the calcium-bound con-G and con-T[K7gamma] dimeric peptides demonstrate a metal-dependent helix-forming motif
J.Am.Chem.Soc., 129, 2007
|
|
7Q5Q
 
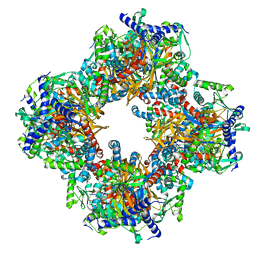 | | Protein community member oxoglutarate dehydrogenase complex E2 core from C. thermophilum | | Descriptor: | Dihydrolipoyllysine-residue succinyltransferase | | Authors: | Chojnowski, G, Skalidis, I, Kyrilis, F.L, Tueting, C, Hamdi, F, Kastritis, P.L. | | Deposit date: | 2021-11-04 | | Release date: | 2022-02-02 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4.38 Å) | | Cite: | Cryo-EM and artificial intelligence visualize endogenous protein community members.
Structure, 30, 2022
|
|
5FYM
 
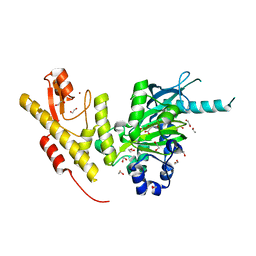 | | Crystal structure of JmjC domain of human histone demethylase UTY in complex with D-2-hydroxyglutarate | | Descriptor: | (2R)-2-hydroxypentanedioic acid, 1,2-ETHANEDIOL, HISTONE DEMETHYLASE UTY, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Gileadi, C, Kupinska, K, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2016-03-08 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Jmjc Domain of Human Histone Demethylase Uty in Complex with D-2- Hydroxyglutarate
To be Published
|
|
4HKH
 
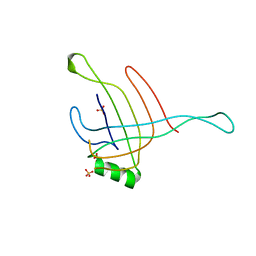 | | Structure of the Hcp1 protein from E. coli EAEC 042 pathovar, mutants N93W-S158W | | Descriptor: | Putative type VI secretion protein, SULFATE ION | | Authors: | Douzi, B, Spinelli, S, Derrez, E, Blangy, S, Brunet, Y.R, Cascales, E, Cambillau, C. | | Deposit date: | 2012-10-15 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure of the Hcp1 protein from E. coli EAEC 042 pathovar, mutants N93W-S158W
To be Published
|
|
8UOZ
 
 | | EmrE structure in the TPP-bound state (WT/E14Q heterodimer) | | Descriptor: | SMR family multidrug efflux protein EmrE, TETRAPHENYLPHOSPHONIUM | | Authors: | Li, J, Sae Her, A, Besch, A, Ramirez, B, Crames, M, Banigan, J.R, Mueller, C, Marsiglia, W.M, Zhang, Y, Traaseth, N.J. | | Deposit date: | 2023-10-20 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | SOLID-STATE NMR, SOLUTION NMR | | Cite: | Dynamics underlie the drug recognition mechanism by the efflux transporter EmrE.
Nat Commun, 15, 2024
|
|
7Q5S
 
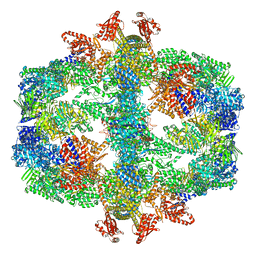 | | Protein community member fatty acid synthase complex from C. thermophilum | | Descriptor: | 3-hydroxyacyl-[acyl-carrier-protein] dehydratase, 3-oxoacyl-[acyl-carrier-protein] reductase | | Authors: | Chojnowski, G, Skalidis, I, Kyrilis, F.L, Tueting, C, Hamdi, F, Kastritis, P.L. | | Deposit date: | 2021-11-04 | | Release date: | 2022-02-02 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4.47 Å) | | Cite: | Cryo-EM and artificial intelligence visualize endogenous protein community members.
Structure, 30, 2022
|
|
5FZ7
 
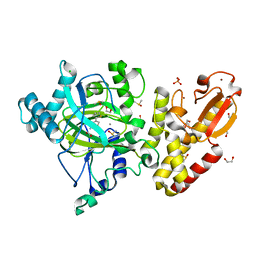 | | Crystal structure of the catalytic domain of human JARID1B in complex with Maybridge fragment ethyl 2-amino-4-thiophen-2-ylthiophene-3- carboxylate (N06131b) (ligand modelled based on PANDDA event map, SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Kupinska, K, Szykowska, A, Pearce, N, Talon, R, Collins, P, Gileadi, C, Strain-Damerell, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, von Delft, F, Brennan, P.E, Oppermann, U. | | Deposit date: | 2016-03-11 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with Maybridge Fragment Ethyl 2-Amino-4-Thiophen-2-Ylthiophene-3-Carboxylate (N06131B) (Ligand Modelled Based on Pandda Event Map, Sgc - Diamond I04-1 Fragment Screening)
To be Published
|
|
4M9C
 
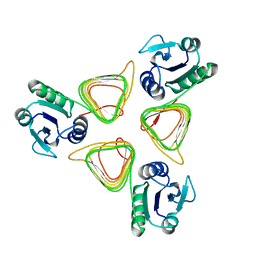 | | WeeI from Acinetobacter baumannii AYE | | Descriptor: | Bacterial transferase hexapeptide (Three repeats) family protein | | Authors: | Morrison, M.J, Imperiali, B. | | Deposit date: | 2013-08-14 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical analysis and structure determination of bacterial acetyltransferases responsible for the biosynthesis of UDP-N,N'-diacetylbacillosamine.
J.Biol.Chem., 288, 2013
|
|
5CCL
 
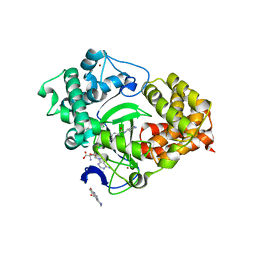 | | Crystal structure of SMYD3 with SAM and oxindole compound | | Descriptor: | 1,2-ETHANEDIOL, 2-oxidanylidene-N-piperidin-4-yl-1,3-dihydroindole-5-carboxamide, Histone-lysine N-methyltransferase SMYD3, ... | | Authors: | Boriack-Sjodin, P.A. | | Deposit date: | 2015-07-02 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Novel Oxindole Sulfonamides and Sulfamides: EPZ031686, the First Orally Bioavailable Small Molecule SMYD3 Inhibitor.
Acs Med.Chem.Lett., 7, 2016
|
|
336D
 
 | | INTERACTION BETWEEN LEFT-HANDED Z-DNA AND POLYAMINE-3 THE CRYSTAL STRUCTURE OF THE D(CG)3 AND THERMOSPERMINE COMPLEX | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), MAGNESIUM ION, N-(3-AMINO-PROPYL)-N-(5-AMINOPROPYL)-1,4-DIAMINOBUTANE | | Authors: | Ohishi, H, Terasoma, N, Nakanishi, I, Van Der Marel, G, Van Boom, J.H, Rich, A, Wang, A.H.-J, Hakoshima, T, Tomita, K.-I. | | Deposit date: | 1997-06-24 | | Release date: | 1998-04-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Interaction between left-handed Z-DNA and polyamine - 3. The crystal structure of the d(CG)3 and thermospermine complex.
FEBS Lett., 398, 1996
|
|
2UVO
 
 | | High Resolution Crystal Structure of Wheat Germ Agglutinin in Complex with N-Acetyl-D-Glucosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, AGGLUTININ ISOLECTIN 1, ... | | Authors: | Schwefel, D, Wittmann, V, Diederichs, K, Welte, W. | | Deposit date: | 2007-03-13 | | Release date: | 2008-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis of Multivalent Binding to Wheat Germ Agglutinin.
J.Am.Chem.Soc., 132, 2010
|
|
8V6Q
 
 | | Crystal structure of EcThsA in ligand-free state | | Descriptor: | Thoeris protein ThsA Macro domain-containing protein | | Authors: | Shi, Y, Masic, V, Mosaiab, T, Goulart, C.C, Hartley-Tassell, L, Sorbello, M, Vasquez, V, Mishra, B.P, Holt, S, Gu, W, Kobe, B, Ve, T. | | Deposit date: | 2023-12-02 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural characterization of macro domain-containing Thoeris antiphage defense systems.
Sci Adv, 10, 2024
|
|
3ABB
 
 | | Crystal structure of CYP105D6 | | Descriptor: | Cytochrome P450 hydroxylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xu, L.H, Fushinobu, S, Takamatsu, S, Wakagi, T, Ikeda, H, Shoun, H. | | Deposit date: | 2009-12-04 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Regio- and stereospecificity of filipin hydroxylation sites revealed by crystal structures of cytochrome P450 105P1 and 105D6 from Streptomyces avermitilis
J.Biol.Chem., 285, 2010
|
|
8V6S
 
 | | Crystal structure of PcThsA in complex with Imidazole Adenine Dinucleotide | | Descriptor: | Thoeris protein ThsA Macro domain-containing protein, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-imidazol-1-yl-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Shi, Y, Masic, V, Mosaiab, T, Goulart, C.C, Hartley-Tassell, L, Sorbello, M, Vasquez, E, Mishra, B.P, Holt, S, Gu, W, Kobe, B, Ve, T. | | Deposit date: | 2023-12-02 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of macro domain-containing Thoeris antiphage defense systems.
Sci Adv, 10, 2024
|
|
8V6R
 
 | | Crystal structure of EcThsA in complex with ADPR | | Descriptor: | Thoeris protein ThsA Macro domain-containing protein, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Shi, Y, Masic, V, Mosaiab, T, Goulart, C.C, Hartley-Tassell, L, Sorbello, M, Vasquez, E, Mishra, B.P, Holt, S, Gu, W, Kobe, B, Ve, T. | | Deposit date: | 2023-12-02 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural characterization of macro domain-containing Thoeris antiphage defense systems.
Sci Adv, 10, 2024
|
|
7L6I
 
 | | The empty AAV13 capsid | | Descriptor: | Capsid protein | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2020-12-23 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Completion of the AAV Structural Atlas: Serotype Capsid Structures Reveals Clade-Specific Features.
Viruses, 13, 2021
|
|
5FDL
 
 | | Crystal Structure of K103N/Y181C Mutant HIV-1 Reverse Transcriptase (RT) in Complex with IDX899 | | Descriptor: | P51 Reverse transcriptase, P66 Reverse transcriptase, methyl (R)-(2-carbamoyl-5-chloro-1H-indol-3-yl)[3-(2-cyanoethyl)-5-methylphenyl]phosphinate | | Authors: | Dousson, C.B, Alexandre, F.-R, Convard, T, Fisher, M, Lamers, M.B.A.C, Leonard, P.M. | | Deposit date: | 2015-12-16 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of the Aryl-phospho-indole IDX899, a Highly Potent Anti-HIV Non-nucleoside Reverse Transcriptase Inhibitor.
J.Med.Chem., 59, 2016
|
|
4HNV
 
 | | Crystal structure of R54E mutant of S. aureus Pyruvate carboxylase | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yu, L.P.C, Tong, L. | | Deposit date: | 2012-10-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterizing the Importance of the Biotin Carboxylase Domain Dimer for Staphylococcus aureus Pyruvate Carboxylase Catalysis.
Biochemistry, 52, 2013
|
|
8UCR
 
 | |
7KGO
 
 | | Crystal Structure of HLA-A*0201in complex with SARS-CoV-2 N351-359 | | Descriptor: | Beta-2-microglobulin, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Szeto, C, Chatzileontiadou, D.S.M, Riboldi-Tunnicliffe, A, Gras, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The presentation of SARS-CoV-2 peptides by the common HLA-A * 02:01 molecule.
Iscience, 24, 2021
|
|
