4G5Q
 
 | | Structure of LGN GL4/Galphai1 complex | | Descriptor: | CITRIC ACID, G-protein-signaling modulator 2, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Jia, M, Li, J, Zhu, J, Wen, W, Zhang, M, Wang, W. | | Deposit date: | 2012-07-18 | | Release date: | 2012-09-05 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of the scaffolding protein LGN reveal the general mechanism by which GoLoco binding motifs inhibit the release of GDP from G alpha i.
J.Biol.Chem., 287, 2012
|
|
3R7M
 
 | | AKR1C3 complex with sulindac | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Yosaatmadja, Y, Teague, R.M, Flanagan, J.U, Squire, C.J. | | Deposit date: | 2011-03-22 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of three classes of non-steroidal anti-inflammatory drugs in complex with aldo-keto reductase 1C3.
Plos One, 7, 2012
|
|
1C0A
 
 | | CRYSTAL STRUCTURE OF THE E. COLI ASPARTYL-TRNA SYNTHETASE : TRNAASP : ASPARTYL-ADENYLATE COMPLEX | | Descriptor: | ADENOSINE MONOPHOSPHATE, ASPARTYL TRNA, ASPARTYL TRNA SYNTHETASE, ... | | Authors: | Eiler, S, Dock-Bregeon, A.-C, Moulinier, L, Thierry, J.-C, Moras, D. | | Deposit date: | 1999-07-15 | | Release date: | 1999-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis of aspartyl-tRNA(Asp) in Escherichia coli--a snapshot of the second step.
EMBO J., 18, 1999
|
|
2X3A
 
 | | AsaP1 inactive mutant E294Q, an extracellular toxic zinc metalloendopeptidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Bogdanovic, X, Palm, G.J, Singh, R.K, Hinrichs, W. | | Deposit date: | 2010-01-22 | | Release date: | 2011-02-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Evidence of Intramolecular Propeptide Inhibition of the Aspzincin Metalloendopeptidase Asap1.
FEBS Lett., 590, 2016
|
|
3WTO
 
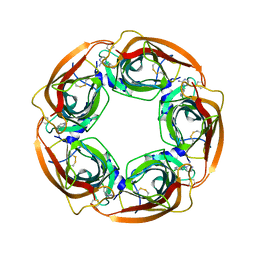 | | Crystal Structure of Lymnaea stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed with Desnitro-imidacloprid | | Descriptor: | (2Z)-1-[(6-chloropyridin-3-yl)methyl]imidazolidin-2-imine, Acetylcholine-binding protein | | Authors: | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Matsuda, K. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Studies on an acetylcholine binding protein identify a basic residue in loop G on the beta 1 strand as a new structural determinant of neonicotinoid actions
Mol.Pharmacol., 86, 2014
|
|
1C10
 
 | | CRYSTAL STRUCTURE OF HEW LYSOZYME UNDER PRESSURE OF XENON (8 BAR) | | Descriptor: | CHLORIDE ION, PROTEIN (LYSOZYME), SODIUM ION, ... | | Authors: | Prange, T, Schiltz, M, Pernot, L, Colloc'h, N, Longhi, S, Bourguet, W, Fourme, R. | | Deposit date: | 1999-07-16 | | Release date: | 1999-07-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Exploring hydrophobic sites in proteins with xenon or krypton.
Proteins, 30, 1998
|
|
4G8D
 
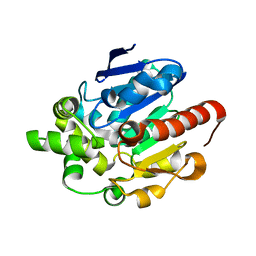 | |
1V7Z
 
 | | creatininase-product complex | | Descriptor: | MANGANESE (II) ION, N-[(E)-AMINO(IMINO)METHYL]-N-METHYLGLYCINE, SULFATE ION, ... | | Authors: | Yoshimoto, T, Tanaka, N, Kanada, N, Inoue, T, Nakajima, Y, Haratake, M, Nakamura, K.T, Xu, Y, Ito, K. | | Deposit date: | 2003-12-26 | | Release date: | 2004-01-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of creatininase reveal the substrate binding site and provide an insight into the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1BPW
 
 | | BETAINE ALDEHYDE DEHYDROGENASE FROM COD LIVER | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTEIN (ALDEHYDE DEHYDROGENASE) | | Authors: | Johansson, K, El Ahmad, M, Ramaswamy, S, Hjelmqvist, L, Jornvall, H, Eklund, H. | | Deposit date: | 1998-08-12 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of betaine aldehyde dehydrogenase at 2.1 A resolution.
Protein Sci., 7, 1998
|
|
4GCD
 
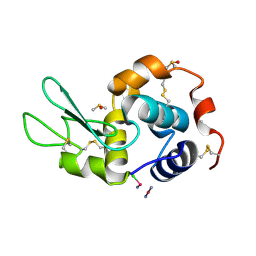 | |
3RHD
 
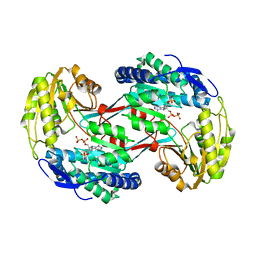 | | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase GapN from Methanocaldococcus jannaschii DSM 2661 complexed with NADP | | Descriptor: | Lactaldehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Malashkevich, V.N, Toro, R, Seidel, R, Garrett, S, Foti, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-04-11 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase GapN from Methanocaldococcus jannaschii DSM 2661 complexed with NADP
To be Published
|
|
3RHH
 
 | | Crystal structure of NADP-dependent glyceraldehyde-3-phosphate dehydrogenase from Bacillus halodurans C-125 complexed with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent glyceraldehyde-3-phosphate dehydrogenase, SULFATE ION | | Authors: | Malashkevich, V.N, Toro, R, Seidel, R, Garrett, S, Foti, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-04-11 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of NADP-dependent glyceraldehyde-3-phosphate dehydrogenase from Bacillus halodurans C-125 complexed with NADP
To be Published
|
|
3O5S
 
 | | Crystal Structure of the endo-beta-1,3-1,4 glucanase from Bacillus subtilis (strain 168) | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucanase, CALCIUM ION | | Authors: | Santos, C.R, Tonoli, C.C.C, Souza, A.R, Furtado, G.P, Ribeiro, L.F, Ward, R.J, Murakami, M.T. | | Deposit date: | 2010-07-28 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and structural characterization of a Beta-1,3 1,4-glucanase from Bacillus subtilis 168
PROCESS BIOCHEM, 46, 2011
|
|
3RI8
 
 | | Xylanase C from Aspergillus kawachii D37N mutant | | Descriptor: | Endo-1,4-beta-xylanase 3 | | Authors: | Fushinobu, S, Uno, T, Kitaoka, M, Hayashi, K, Matsuzawa, H, Wakagi, T. | | Deposit date: | 2011-04-13 | | Release date: | 2011-10-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutational analysis of fungal family 11 xylanases on pH optimum determination
J.APPL.GLYOSCI., 58, 2011
|
|
3RIL
 
 | | The acid beta-glucosidase active site exhibits plasticity in binding 3,4,5,6-tetrahydroxyazepane-based inhibitors: implications for pharmacological chaperone design for gaucher disease | | Descriptor: | (3S,4R,5R,6S)-azepane-3,4,5,6-tetrol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glucosylceramidase, ... | | Authors: | Orwig, S.D, Lieberman, R.L. | | Deposit date: | 2011-04-13 | | Release date: | 2012-03-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Binding of 3,4,5,6-tetrahydroxyazepanes to the acid-beta-glucosidase active site: implications for pharmacological chaperone design for Gaucher disease
Biochemistry, 50, 2011
|
|
4GGP
 
 | |
3RIT
 
 | | Crystal structure of Dipeptide Epimerase from Methylococcus capsulatus complexed with Mg and dipeptide L-Arg-D-Lys | | Descriptor: | ARGININE, D-LYSINE, Dipeptide epimerase, ... | | Authors: | Lukk, T, Sakai, A, Song, L, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2011-04-14 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1UMB
 
 | | branched-chain 2-oxo acid dehydrogenase (E1) from Thermus thermophilus HB8 in holo-form | | Descriptor: | 2-oxo acid dehydrogenase alpha subunit, 2-oxo acid dehydrogenase beta subunit, MAGNESIUM ION, ... | | Authors: | Nakai, T, Nakagawa, N, Maoka, N, Masui, R, Kuramitsu, S, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-25 | | Release date: | 2004-03-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ligand-induced Conformational Changes and a Reaction Intermediate in Branched-chain 2-Oxo Acid Dehydrogenase (E1) from Thermus thermophilus HB8, as Revealed by X-ray Crystallography
J.Mol.Biol., 337, 2004
|
|
4GIE
 
 | |
1V5W
 
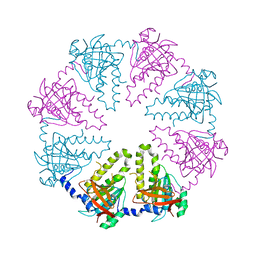 | | Crystal structure of the human Dmc1 protein | | Descriptor: | Meiotic recombination protein DMC1/LIM15 homolog | | Authors: | Kinebuchi, T, Kagawa, W, Enomoto, R, Ikawa, S, Shibata, T, Kurumizaka, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-26 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for octameric ring formation and DNA interaction of the human homologous-pairing protein dmc1
Mol.Cell, 14, 2004
|
|
1UR2
 
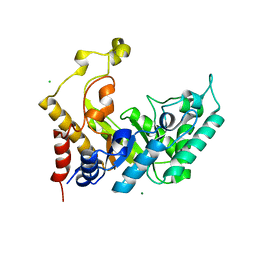 | | Xylanase Xyn10B mutant (E262S) from Cellvibrio mixtus in complex with arabinofuranose alpha 1,3 linked to xylotriose | | Descriptor: | CHLORIDE ION, ENDOXYLANASE, MAGNESIUM ION, ... | | Authors: | Pell, G, Taylor, E.J, Gloster, T.M, Turkenburg, J.P, Fontes, C.M.G.A, Ferreira, L.M.A, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2003-10-24 | | Release date: | 2003-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Mechanisms by which Family 10 Glycoside Hydrolases Bind Decorated Substrates
J.Biol.Chem., 279, 2004
|
|
3RJY
 
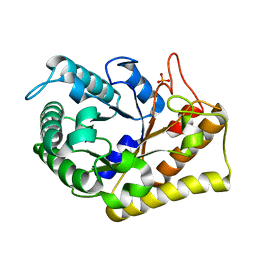 | | Crystal Structure of Hyperthermophilic Endo-beta-1,4-glucanase in complex with substrate | | Descriptor: | Endoglucanase FnCel5A, PHOSPHATE ION, alpha-D-glucopyranose | | Authors: | Zheng, B, Yang, W, Zhao, X, Wang, Y, Lou, Z, Rao, Z, Feng, Y. | | Deposit date: | 2011-04-15 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of hyperthermophilic Endo-beta-1,4-glucanase: Implications for catalytic mechanism and thermostability.
J.Biol.Chem., 287, 2012
|
|
4GIJ
 
 | | Crystal Structure of Pseudouridine Monophosphate Glycosidase Complexed with Sulfate | | Descriptor: | MANGANESE (II) ION, Pseudouridine-5'-phosphate glycosidase, SULFATE ION | | Authors: | Huang, S, Mahanta, N, Begley, T.P, Ealick, S.E. | | Deposit date: | 2012-08-08 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.941 Å) | | Cite: | Pseudouridine monophosphate glycosidase: a new glycosidase mechanism.
Biochemistry, 51, 2012
|
|
1BSV
 
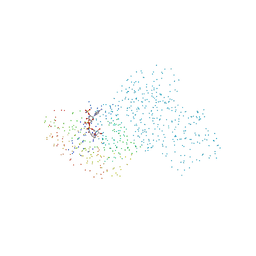 | | GDP-FUCOSE SYNTHETASE FROM ESCHERICHIA COLI COMPLEX WITH NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTEIN (GDP-FUCOSE SYNTHETASE) | | Authors: | Somers, W.S, Stahl, M.L, Sullivan, F.X. | | Deposit date: | 1998-08-31 | | Release date: | 1999-08-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | GDP-fucose synthetase from Escherichia coli: structure of a unique member of the short-chain dehydrogenase/reductase family that catalyzes two distinct reactions at the same active site.
Structure, 6, 1998
|
|
3ROS
 
 | |
