1G92
 
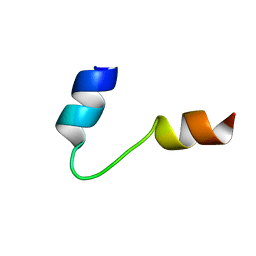 | | SOLUTION STRUCTURE OF PONERATOXIN | | Descriptor: | PONERATOXIN | | Authors: | Szolajska, E, Poznanski, J, Ferber, M.L, Michalik, J, Gout, E, Fender, P, Bailly, I, Dublet, B, Chroboczek, J. | | Deposit date: | 2000-11-22 | | Release date: | 2003-11-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Poneratoxin, a neurotoxin from ant venom. Structure and expression in insect cells and construction of a bio-insecticide.
Eur.J.Biochem., 271, 2004
|
|
1JPU
 
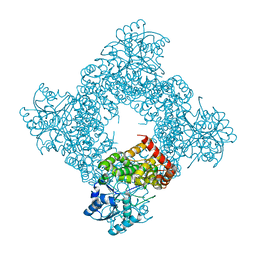 | | Crystal Structure of Bacillus Stearothermophilus Glycerol Dehydrogenase | | Descriptor: | ZINC ION, glycerol dehydrogenase | | Authors: | Ruzheinikov, S.N, Burke, J, Sedelnikova, S, Baker, P.J, Taylor, R, Bullough, P.A, Muir, N.M, Gore, M.G, Rice, D.W. | | Deposit date: | 2001-08-03 | | Release date: | 2001-10-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glycerol dehydrogenase. structure, specificity, and mechanism of a family III polyol dehydrogenase.
Structure, 9, 2001
|
|
1GAZ
 
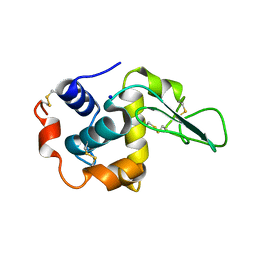 | | Crystal Structure of Mutant Human Lysozyme Substituted at the Surface Positions | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-06-26 | | Release date: | 2000-07-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of amino acid residues at turns in the conformational stability and folding of human lysozyme.
Biochemistry, 39, 2000
|
|
1GB5
 
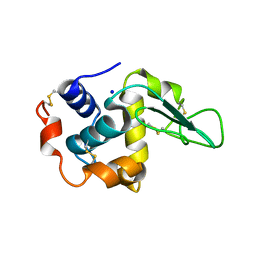 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-06-26 | | Release date: | 2000-07-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of surface hydrophobic residues in the conformational stability of human lysozyme at three different positions.
Biochemistry, 39, 2000
|
|
6G8S
 
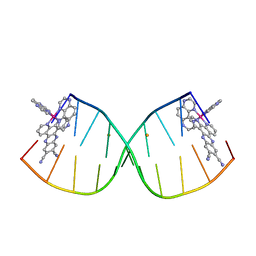 | | [Ru(TAP)2(11,12-CN2-dppz)]2+ bound to d(CCGGACCCGG/CCGGGTCCGG)2 | | Descriptor: | BARIUM ION, DNA (5'-D(*CP*CP*GP*GP*AP*CP*CP*CP*GP*G)-3'), DNA (5'-D(*CP*CP*GP*GP*GP*TP*CP*CP*GP*G)-3'), ... | | Authors: | McQuaid, K.T, Hall, J.P, Cardin, C.J. | | Deposit date: | 2018-04-09 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | X-ray Crystal Structures Show DNA Stacking Advantage of Terminal Nitrile Substitution in Ru-dppz Complexes.
Chemistry, 24, 2018
|
|
1FZ7
 
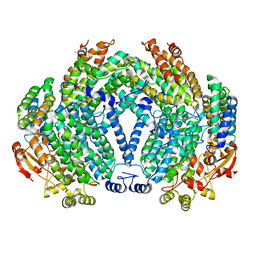 | | METHANE MONOOXYGENASE HYDROXYLASE, FORM III SOAKED IN 0.9 M ETHANOL | | Descriptor: | CALCIUM ION, ETHANOL, FE (III) ION, ... | | Authors: | Whittington, D.A, Sazinsky, M.H, Lippard, S.J. | | Deposit date: | 2000-10-03 | | Release date: | 2001-02-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | X-ray crystal structure of alcohol products bound at the active site of soluble methane monooxygenase hydroxylase.
J.Am.Chem.Soc., 123, 2001
|
|
1KF6
 
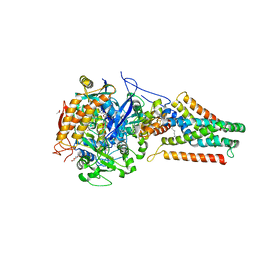 | | E. coli Quinol-Fumarate Reductase with Bound Inhibitor HQNO | | Descriptor: | 2-HEPTYL-4-HYDROXY QUINOLINE N-OXIDE, ACETATE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Iverson, T.M, Luna-Chavez, C, Croal, L.R, Cecchini, G, Rees, D.C. | | Deposit date: | 2001-11-19 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic studies of the Escherichia coli quinol-fumarate reductase with inhibitors bound to the
quinol-binding site.
J.Biol.Chem., 277, 2002
|
|
1GEJ
 
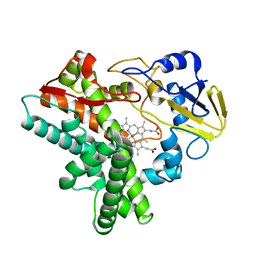 | | STRUCTURAL CHARACTERIZATION OF N-BUTYL-ISOCYANIDE COMPLEXES OF CYTOCHROMES P450NOR AND P450CAM | | Descriptor: | CYTOCHROME P450 55A1, N-BUTYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | lee, D.-S, Park, S.-Y, Yamane, K, Shiro, Y. | | Deposit date: | 2000-11-13 | | Release date: | 2000-12-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural characterization of n-butyl-isocyanide complexes of cytochromes P450nor and P450cam.
Biochemistry, 40, 2001
|
|
1GF0
 
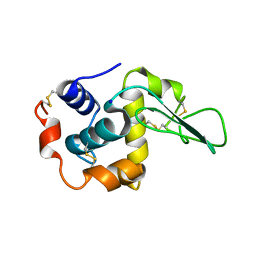 | | BURIED POLAR MUTANT HUMAN LYSOZYME | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-11-30 | | Release date: | 2001-04-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of polar groups in the interior of a protein to the conformational stability.
Biochemistry, 40, 2001
|
|
4KBM
 
 | |
1GF6
 
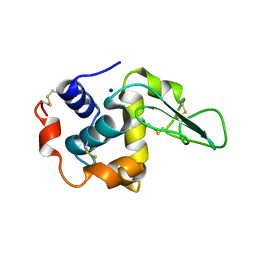 | | BURIED POLAR MUTANT HUMAN LYSOZYME | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-11-30 | | Release date: | 2001-04-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of polar groups in the interior of a protein to the conformational stability.
Biochemistry, 40, 2001
|
|
3J2U
 
 | | Kinesin-13 KLP10A HD in complex with CS-tubulin and a microtubule | | Descriptor: | Kinesin-like protein Klp10A, Tubulin alpha-1A chain, Tubulin beta-2B chain | | Authors: | Asenjo, A.B, Chatterjee, C, Tan, D, DePaoli, V, Rice, W.J, Diaz-Avalos, R, Silvestry, M, Sosa, H. | | Deposit date: | 2013-01-10 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (10.8 Å) | | Cite: | Structural model for tubulin recognition and deformation by Kinesin-13 microtubule depolymerases.
Cell Rep, 3, 2013
|
|
7WVK
 
 | | Crystal structure of human WDR5 in complex with compound 19 | | Descriptor: | 1,2-ETHANEDIOL, 1-[2,5-bis(chloranyl)phenyl]sulfonylbenzimidazole, GLYCEROL, ... | | Authors: | Han, Q.L, Zhang, X.L, Wang, L, Ren, P.X, Cao, Y, Li, K, Bai, F. | | Deposit date: | 2022-02-10 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Discovery, evaluation and mechanism study of WDR5-targeted small molecular inhibitors for neuroblastoma.
Acta Pharmacol.Sin., 44, 2023
|
|
1GFE
 
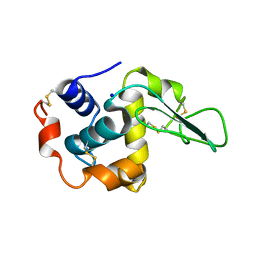 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-12-04 | | Release date: | 2000-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1G0G
 
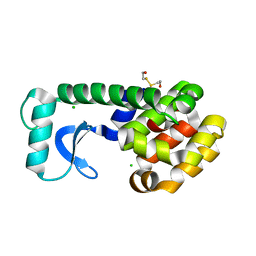 | | CRYSTAL STRUCTURE OF T4 LYSOZYME MUTANT T152A | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | Authors: | Xu, J, Baase, W.A, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2000-10-06 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
1KG2
 
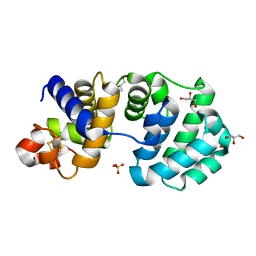 | | Crystal structure of the core fragment of MutY from E.coli at 1.2A resolution | | Descriptor: | A/G-specific adenine glycosylase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Gilboa, R, Kilshtein, A, Zharkov, D.O, Kycia, J.H, Gerchman, S.E, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-11-26 | | Release date: | 2002-11-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Analysis of the E.coli MutY DNA glycosylase structure and function by site-directed mutagenesis
To be Published
|
|
1G0K
 
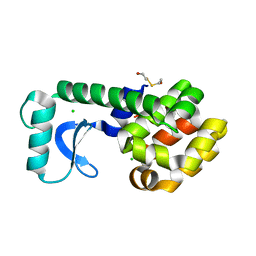 | | CRYSTAL STRUCTURE OF T4 LYSOZYME MUTANT T152C | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | Authors: | Xu, J, Baase, W.A, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2000-10-06 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
1ZDE
 
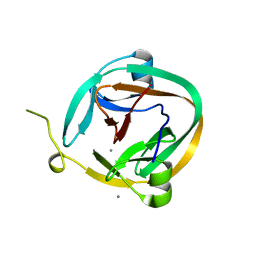 | | 1.95 Angstrom Crystal Structure of a dnaE Intein Precursor from Synechocystis Sp. Pcc 6803 | | Descriptor: | CALCIUM ION, DNA polymerase III alpha subunit, ZINC ION | | Authors: | Sun, P, Ye, S, Ferrandon, S, Evans, T.C, Xu, M.Q, Rao, Z. | | Deposit date: | 2005-04-14 | | Release date: | 2006-01-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of an intein from the split dnaE gene of Synechocystis sp. PCC6803 reveal the catalytic model without the penultimate histidine and the mechanism of zinc ion inhibition of protein splicing
J.Mol.Biol., 353, 2005
|
|
2AS6
 
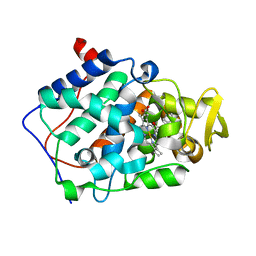 | | cytochrome c peroxidase in complex with cyclopentylamine | | Descriptor: | CYCLOPENTANAMINE, Cytochrome c peroxidase, mitochondrial, ... | | Authors: | Brenk, R, Vetter, S.W, Boyce, S.E, Goodin, D.B, Shoichet, B.K. | | Deposit date: | 2005-08-22 | | Release date: | 2006-04-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Probing molecular docking in a charged model binding site.
J.Mol.Biol., 357, 2006
|
|
1G19
 
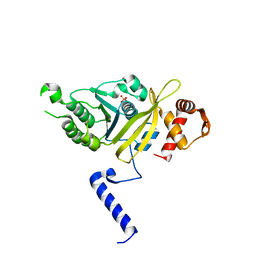 | | STRUCTURE OF RECA PROTEIN | | Descriptor: | PHOSPHATE ION, RECA PROTEIN | | Authors: | Datta, S, Prabu, M.M, Vaze, M.B, Ganesh, N, Chandra, N.R, Muniyappa, K, Vijayan, M, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2000-10-11 | | Release date: | 2000-12-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis RecA and its complex with ADP-AlF(4): implications for decreased ATPase activity and molecular aggregation
Nucleic Acids Res., 28, 2000
|
|
1KSC
 
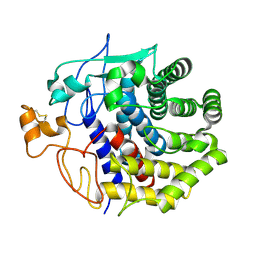 | | The structure of Endoglucanase from termite, Nasutitermes takasagoensis, at pH 5.6. | | Descriptor: | CALCIUM ION, Endo-b-1,4-glucanase | | Authors: | Khademi, S, Guarino, L.A, Watanabe, H, Tokuda, G, Meyer, E.F. | | Deposit date: | 2002-01-11 | | Release date: | 2003-01-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of an endoglucanase from termite, Nasutitermes takasagoensis.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
3AU5
 
 | |
6HN2
 
 | | Ternary complex of Estrogen Receptor alpha peptide and 14-3-3 sigma C42 mutant bound to disulfide fragment PPI stabilizer 5 | | Descriptor: | 14-3-3 protein sigma, 2-(3,4-dichlorophenyl)-~{N}-(2-sulfanylethyl)ethanamide, Estrogen Receptor, ... | | Authors: | Sijbesma, E, Hallenbeck, K.K, Leysen, S, Arkin, M.R, Ottmann, C. | | Deposit date: | 2018-09-13 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Site-Directed Fragment-Based Screening for the Discovery of Protein-Protein Interaction Stabilizers.
J. Am. Chem. Soc., 141, 2019
|
|
2ASS
 
 | | Crystal structure of the Skp1-Skp2-Cks1 complex | | Descriptor: | BENZAMIDINE, Cyclin-dependent kinases regulatory subunit 1, PHOSPHATE ION, ... | | Authors: | Hao, B, Zhang, N, Schulman, B.A, Wu, G, Pagano, M, Pavletich, N.P. | | Deposit date: | 2005-08-24 | | Release date: | 2005-10-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of the Cks1-Dependent Recognition of p27(Kip1) by the SCF(Skp2) Ubiquitin Ligase.
Mol.Cell, 20, 2005
|
|
1KG7
 
 | | Crystal Structure of the E161A mutant of E.coli MutY (core fragment) | | Descriptor: | A/G-specific adenine glycosylase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Gilboa, R, Kilshtein, A, Zharkov, D.O, Kycia, J.H, Gerchman, S.E, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-11-26 | | Release date: | 2002-11-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Analysis of the E.coli MutY DNA glycosylase structure and function by site-directed mutagenesis
To be Published
|
|
