8I1Z
 
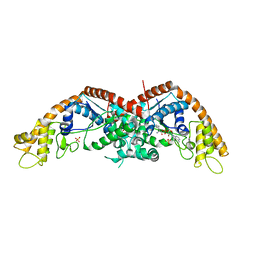 | | E. coli tryptophanyl-tRNA synthetase bound with a chemical fragment | | Descriptor: | 1-(2,3-dihydro-1-benzofuran-5-yl)ethanone, SULFATE ION, TRYPTOPHANYL-5'AMP, ... | | Authors: | Xiang, M, Zhou, H. | | Deposit date: | 2023-01-13 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An asymmetric structure of bacterial TrpRS supports the half-of-the-sites catalytic mechanism and facilitates antimicrobial screening.
Nucleic Acids Res., 51, 2023
|
|
1SZ1
 
 | |
6NY2
 
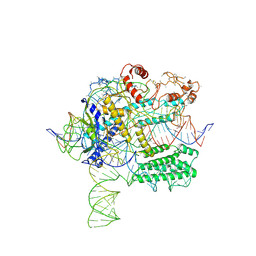 | | CasX-gRNA-DNA(45bp) state I | | Descriptor: | CasX, DNA Non-target strand, DNA target strand, ... | | Authors: | Liu, J.J, Orlova, N, Nogales, E, Doudna, J.A. | | Deposit date: | 2019-02-10 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | CasX enzymes comprise a distinct family of RNA-guided genome editors.
Nature, 566, 2019
|
|
6NY1
 
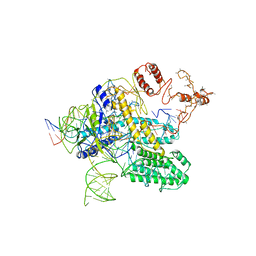 | | CasX-gRNA-DNA(30bp) State II | | Descriptor: | CasX, DNA Non-target strand, DNA target strand, ... | | Authors: | Liu, J.J, Orlova, N, Nogales, E, Doudna, J.A. | | Deposit date: | 2019-02-10 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | CasX enzymes comprise a distinct family of RNA-guided genome editors.
Nature, 566, 2019
|
|
4AYB
 
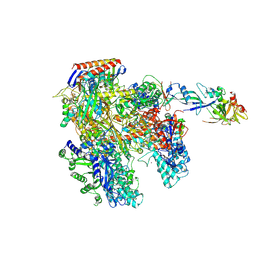 | | RNAP at 3.2Ang | | Descriptor: | DNA-DIRECTED RNA POLYMERASE, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Wojtas, M.N, Mogni, M, Millet, O, Bell, S.D, Abrescia, N.G.A. | | Deposit date: | 2012-06-19 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Structural and Functional Analyses of the Interaction of Archaeal RNA Polymerase with DNA.
Nucleic Acids Res., 40, 2012
|
|
7WAZ
 
 | |
8H99
 
 | | Crystal structure of E. coli ThrS catalytic domain mutant | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, IMIDAZOLE, MAGNESIUM ION, ... | | Authors: | Qiao, H, Xia, M, Wang, J, Fang, P. | | Deposit date: | 2022-10-25 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Tyrosine-targeted covalent inhibition of a tRNA synthetase aided by zinc ion.
Commun Biol, 6, 2023
|
|
8H9C
 
 | | Crystal structure of chemically modified E. coli ThrS catalytic domain 4 | | Descriptor: | N-(2,3-dihydroxybenzoyl)-4-(4-nitrophenyl)-L-threonine, Threonine--tRNA ligase, ZINC ION | | Authors: | Qiao, H, Xia, M, Wang, J, Fang, P. | | Deposit date: | 2022-10-25 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Tyrosine-targeted covalent inhibition of a tRNA synthetase aided by zinc ion.
Commun Biol, 6, 2023
|
|
8H9B
 
 | | Crystal structure of chemically modified E. coli ThrS catalytic domain 3 | | Descriptor: | N-(2,3-dihydroxybenzoyl)-4-(4-nitrophenyl)-L-threonine, Threonine--tRNA ligase, ZINC ION | | Authors: | Qiao, H, Xia, M, Wang, J, Fang, P. | | Deposit date: | 2022-10-25 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Tyrosine-targeted covalent inhibition of a tRNA synthetase aided by zinc ion.
Commun Biol, 6, 2023
|
|
8H9A
 
 | | Crystal structure of chemically modified E. coli ThrS catalytic domain 2 | | Descriptor: | N-(2,3-dihydroxybenzoyl)-4-(4-nitrophenyl)-L-threonine, Threonine--tRNA ligase, ZINC ION | | Authors: | Qiao, H, Xia, M, Wang, J, Fang, P. | | Deposit date: | 2022-10-25 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tyrosine-targeted covalent inhibition of a tRNA synthetase aided by zinc ion.
Commun Biol, 6, 2023
|
|
9H82
 
 | | Small circular RNA dimer - Class 1 | | Descriptor: | RNA (36-MER), RNA (5'-R(P*AP*UP*CP*UP*UP*CP*UP*UP*CP*GP*AP*UP*UP*UP*CP*UP*UP*CP*CP*AP*UP*C)-3') | | Authors: | McRae, E.K, Kristoffersen, E.L, Holliger, P, Andersen, E.S. | | Deposit date: | 2024-10-28 | | Release date: | 2024-12-04 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (7.29 Å) | | Cite: | Roles of dimeric intermediates in RNA-catalyzed rolling circle synthesis.
Nucleic Acids Res., 53, 2025
|
|
8S6W
 
 | |
7WAY
 
 | |
5I9D
 
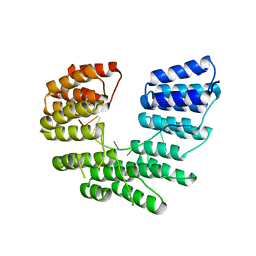 | | Crystal structure of designed pentatricopeptide repeat protein dPPR-U8A2 in complex with its target RNA U8A2 | | Descriptor: | RNA (5'-R(*GP*GP*GP*G*UP*UP*UP*UP*AP*AP*UP*UP*UP*UP*CP*CP*CP*C)-3'), pentatricopeptide repeat protein dPPR-U8A2 | | Authors: | Shen, C, Zhang, D, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-02-20 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins.
Nat Commun, 7, 2016
|
|
5I9G
 
 | | Crystal structure of designed pentatricopeptide repeat protein dPPR-U8C2 in complex with its target RNA U8C2 | | Descriptor: | RNA (5'-R(*GP*GP*G*GP*UP*UP*UP*UP*CP*CP*UP*UP*UP*UP*CP*CP*CP*C)-3'), pentatricopeptide repeat protein dPPR-U8C2 | | Authors: | Shen, C, Zhang, D, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-02-20 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.288 Å) | | Cite: | Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins.
Nat Commun, 7, 2016
|
|
5I9F
 
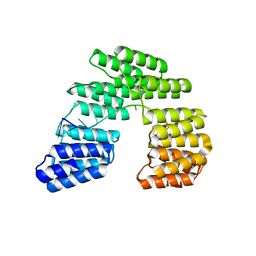 | | Crystal structure of designed pentatricopeptide repeat protein dPPR-U10 in complex with its target RNA U10 | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*CP*CP*CP*C)-3'), pentatricopeptide repeat protein dPPR-U10 | | Authors: | Shen, C, Zhang, D, Guan, Z, Zou, T, Yin, Y. | | Deposit date: | 2016-02-20 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins.
Nat Commun, 7, 2016
|
|
5I9H
 
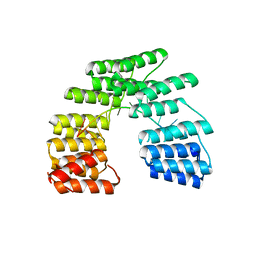 | | Crystal structure of designed pentatricopeptide repeat protein dPPR-U8G2 in complex with its target RNA U8G2 | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*UP*UP*UP*UP*GP*GP*UP*UP*UP*UP*CP*CP*CP*C)-3'), pentatricopeptide repeat protein dPPR-U8G2 | | Authors: | Shen, C, Zhang, D, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-02-20 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins.
Nat Commun, 7, 2016
|
|
1BGZ
 
 | |
9CPJ
 
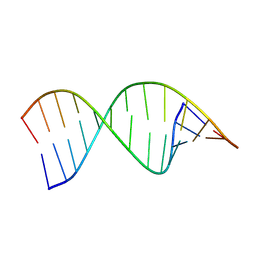 | | Structures of small molecules bound to RNA repeat expansions that cause Huntington's disease-like 2 and myotonic dystrophy type 1 | | Descriptor: | RNA (5'-R(*GP*AP*CP*AP*GP*CP*UP*GP*CP*UP*GP*UP*C)-3') | | Authors: | Chen, J.L, Taghavi, A, Disney, M.D, Fountain, M.A, Childs-Disney, J.L. | | Deposit date: | 2024-07-18 | | Release date: | 2024-08-07 | | Method: | SOLUTION NMR | | Cite: | Structures of small molecules bound to RNA repeat expansions that cause Huntington's disease-like 2 and myotonic dystrophy type 1.
Bioorg.Med.Chem.Lett., 2024
|
|
7WB0
 
 | |
8XMC
 
 | | Post-translocated Pol IV transcription elongation complex | | Descriptor: | DNA-directed RNA polymerase IV subunit 1, DNA-directed RNA polymerase IV subunit 7, DNA-directed RNA polymerases II and IV subunit 5A, ... | | Authors: | Huang, K, Fang, C.L, Zhang, Y. | | Deposit date: | 2023-12-27 | | Release date: | 2024-07-24 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Transcription elongation of the plant RNA polymerase IV is prone to backtracking.
Sci Adv, 10, 2024
|
|
8XMB
 
 | | NTP-bound Pol IV transcription elongation complex | | Descriptor: | DNA-directed RNA polymerase IV subunit 1, DNA-directed RNA polymerase IV subunit 7, DNA-directed RNA polymerases II and IV subunit 5A, ... | | Authors: | Huang, K, Fang, C.L, Zhang, Y. | | Deposit date: | 2023-12-27 | | Release date: | 2024-07-24 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Transcription elongation of the plant RNA polymerase IV is prone to backtracking.
Sci Adv, 10, 2024
|
|
8XMD
 
 | | Pre-translocated Pol IV transcription elongation complex | | Descriptor: | DNA-directed RNA polymerase IV subunit 1, DNA-directed RNA polymerase IV subunit 7, DNA-directed RNA polymerases II and IV subunit 5A, ... | | Authors: | Huang, K, Fang, C.L, Zhang, Y. | | Deposit date: | 2023-12-27 | | Release date: | 2024-07-24 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Transcription elongation of the plant RNA polymerase IV is prone to backtracking.
Sci Adv, 10, 2024
|
|
8XME
 
 | | Backtracked Pol IV transcription elongation complex | | Descriptor: | DNA-directed RNA polymerase IV subunit 1, DNA-directed RNA polymerase IV subunit 7, DNA-directed RNA polymerases II and IV subunit 5A, ... | | Authors: | Huang, K, Fang, C.L, Zhang, Y. | | Deposit date: | 2023-12-27 | | Release date: | 2024-07-24 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Transcription elongation of the plant RNA polymerase IV is prone to backtracking.
Sci Adv, 10, 2024
|
|
6MWN
 
 | |
