1CIO
 
 | | RECOMBINANT SPERM WHALE MYOGLOBIN I99V MUTANT (MET) | | Descriptor: | PROTEIN (MYOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Liong, E.C, Phillips Jr, G.N. | | Deposit date: | 1999-04-01 | | Release date: | 1999-04-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Waterproofing the heme pocket. Role of proximal amino acid side chains in preventing hemin loss from myoglobin.
J.Biol.Chem., 276, 2001
|
|
4YWU
 
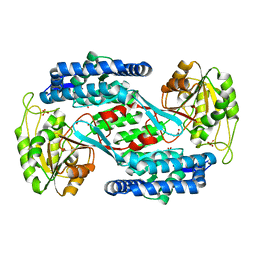 | | Structural insight into the substrate inhibition mechanism of NADP+-dependent succinic semialdehyde dehydrogenase from Streptococcus pyogenes | | Descriptor: | 4-oxobutanoic acid, SULFATE ION, Succinic semialdehyde dehydrogenase | | Authors: | Jang, E.H, Park, S.A, Chi, Y.M, Lee, K.S. | | Deposit date: | 2015-03-21 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insight into the substrate inhibition mechanism of NADP(+)-dependent succinic semialdehyde dehydrogenase from Streptococcus pyogenes.
Biochem.Biophys.Res.Commun., 461, 2015
|
|
5DUD
 
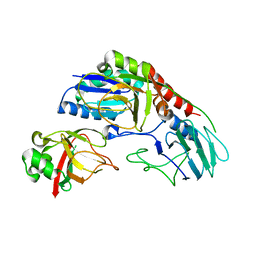 | | Crystal structure of E. coli YbgJK | | Descriptor: | YbgJ, YbgK | | Authors: | Arbing, M.A, Kaufmann, M, Shin, A, Medrano-Soto, A, Cascio, D, Eisenberg, D. | | Deposit date: | 2015-09-18 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of E. coli YbgJK
To Be Published
|
|
2CMB
 
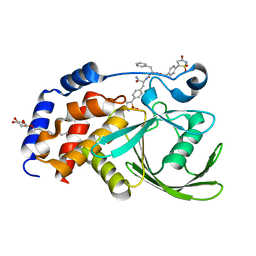 | | Structural Basis for Inhibition of Protein Tyrosine Phosphatase 1B by Isothiazolidinone Heterocyclic Phosphonate Mimetics | | Descriptor: | N-{[4-(1,1-DIOXIDO-3-OXO-2,3-DIHYDROISOTHIAZOL-5-YL)PHENYL]ACETYL}-L-PHENYLALANYL-4-(1,1-DIOXIDO-3-OXO-2,3-DIHYDROISOTHIAZOL-5-YL)-L-PHENYLALANINAMIDE, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1, octyl beta-D-glucopyranoside | | Authors: | Ala, P.J, Gonneville, L, Hillman, M.C, Becker-Pasha, M, Wei, M, Reid, B.G, Klabe, R, Yue, E.W, Wayland, B, Douty, B, Combs, A.P, Polam, P, Wasserman, Z, Bower, M, Burn, T.C, Hollis, G.F, Wynn, R. | | Deposit date: | 2006-05-04 | | Release date: | 2006-08-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Inhibition of Protein-Tyrosine Phosphatase 1B by Isothiazolidinone Heterocyclic Phosphonate Mimetics.
J.Biol.Chem., 281, 2006
|
|
2X7F
 
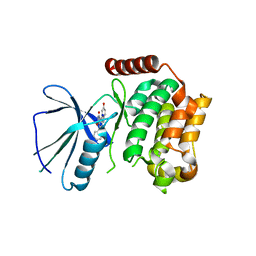 | | Crystal structure of the kinase domain of human Traf2- and Nck- interacting Kinase with Wee1Chk1 inhibitor | | Descriptor: | 9-HYDROXY-4-PHENYLPYRROLO[3,4-C]CARBAZOLE-1,3(2H,6H)-DIONE, SODIUM ION, TRAF2 AND NCK-INTERACTING PROTEIN KINASE | | Authors: | Vollmar, M, Alfano, I, Shrestha, B, Bray, J, Muniz, J.R.C, Roos, A, Filippakopoulos, P, Burgess-Brown, N, Ugochukwu, E, Gileadi, O, Phillips, C, Mahajan, P, Pike, A.C.W, Fedorov, O, Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S. | | Deposit date: | 2010-02-26 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Kinase Domain of Human Traf2- and Nck-Interacting Kinase with Wee1Chk1 Inhibitor
To be Published
|
|
1HOR
 
 | | STRUCTURE AND CATALYTIC MECHANISM OF GLUCOSAMINE 6-PHOSPHATE DEAMINASE FROM ESCHERICHIA COLI AT 2.1 ANGSTROMS RESOLUTION | | Descriptor: | 2-DEOXY-2-AMINO GLUCITOL-6-PHOSPHATE, GLUCOSAMINE 6-PHOSPHATE DEAMINASE, PHOSPHATE ION | | Authors: | Oliva, G, Fontes, M.R.M, Garratt, R.C, Altamirano, M.M, Calcagno, M.L, Horjales, E. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and catalytic mechanism of glucosamine 6-phosphate deaminase from Escherichia coli at 2.1 A resolution.
Structure, 3, 1995
|
|
1O8B
 
 | | Structure of Escherichia coli ribose-5-phosphate isomerase, RpiA, complexed with arabinose-5-phosphate. | | Descriptor: | 5-O-phosphono-beta-D-arabinofuranose, RIBOSE 5-PHOSPHATE ISOMERASE | | Authors: | Zhang, R.-g, Andersson, C.E, Savchenko, A, Skarina, T, Evdokimova, E, Beasley, S, Arrowsmith, C.H, Edwards, A.M, Joachimiak, A, Mowbray, S.L, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-11-26 | | Release date: | 2003-01-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure of Escherichia Coli Ribose-5-Phosphate Isomerase: A Ubiquitous Enzyme of the Pentose Phosphate Pathway and the Calvin Cycle
Structure, 11, 2003
|
|
6HOJ
 
 | | Structure of Beclin1 LIR motif bound to GABARAP | | Descriptor: | 1,2-ETHANEDIOL, Beclin-1,Gamma-aminobutyric acid receptor-associated protein, SULFATE ION | | Authors: | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
2VKG
 
 | | COMPLEXES OF DODECIN WITH FLAVIN AND FLAVIN-LIKE LIGANDS | | Descriptor: | CHLORIDE ION, DODECIN, MAGNESIUM ION, ... | | Authors: | Grininger, M, Noell, G, Trawoeger, S, Sinner, E, Oesterhelt, D. | | Deposit date: | 2007-12-19 | | Release date: | 2008-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Electrochemical Switching of the Flavoprotein Dodecin at Gold Surfaces Modified by Flavin-DNA Hybrid Linkers
Biointerphases, 3, 2008
|
|
6HPF
 
 | | Structure of Inactive E165Q mutant of fungal non-CBM carrying GH26 endo-b-mannanase from Yunnania penicillata in complex with alpha-62-61-di-galactosyl-mannotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, CHLORIDE ION, ... | | Authors: | von Freiesleben, P, Moroz, O.V, Blagova, E, Wiemann, M, Spodsberg, N, Agger, J.W, Davies, G.J, Wilson, K.S, Stalbrand, H, Meyer, A.S, Krogh, K.B.R.M. | | Deposit date: | 2018-09-20 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal structure and substrate interactions of an unusual fungal non-CBM carrying GH26 endo-beta-mannanase from Yunnania penicillata.
Sci Rep, 9, 2019
|
|
7L4E
 
 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase Ketosynthase, FabF, and C16:1-crypto Acyl Carrier Protein, AcpP | | Descriptor: | 1,2-ETHANEDIOL, 3-oxoacyl-[acyl-carrier-protein] synthase 2, Acyl carrier protein, ... | | Authors: | Mindrebo, J.T, Chen, A, Kim, W.E, Burkart, M.D, Noel, J.P. | | Deposit date: | 2020-12-18 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Mechanistic Analyses of the Gating Mechanism of Elongating Ketosynthases
Acs Catalysis, 11, 2021
|
|
6DTM
 
 | | Crystal Structure of Helicobacter pylori TlpA Chemoreceptor Ligand Binding Domain | | Descriptor: | CHLORIDE ION, Methyl-accepting chemotaxis protein TlpA | | Authors: | Remington, S.J, Guillemin, K, Sweeney, E, Perkins, A. | | Deposit date: | 2018-06-17 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the ligand-binding domain of Helicobacter pylori chemoreceptor TlpA.
Protein Sci., 27, 2018
|
|
2J0S
 
 | | The crystal structure of the Exon Junction Complex at 2.2 A resolution | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP *UP*UP*UP*UP*U)-3', ATP-DEPENDENT RNA HELICASE DDX48, MAGNESIUM ION, ... | | Authors: | Bono, F, Ebert, J, Lorentzen, E, Conti, E. | | Deposit date: | 2006-08-04 | | Release date: | 2006-09-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The Crystal Structure of the Exon Junction Complex Reveals How It Mantains a Stable Grip on Mrna
Cell(Cambridge,Mass.), 126, 2006
|
|
2O30
 
 | | Nuclear movement protein from E. cuniculi GB-M1 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, NUCLEAR MOVEMENT PROTEIN | | Authors: | Binkowski, T.A, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-11-30 | | Release date: | 2007-01-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Nuclear movement protein from E. cuniculi GB-M1
TO BE PUBLISHED
|
|
6HT1
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with SGC-iMLLT (compound 92) | | Descriptor: | 1,2-ETHANEDIOL, 1-methyl-~{N}-[2-[[(2~{S})-2-methylpyrrolidin-1-yl]methyl]-3~{H}-benzimidazol-5-yl]indazole-5-carboxamide, Protein ENL, ... | | Authors: | Heidenreich, D, Chaikuad, A, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-10-02 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of an MLLT1/3 YEATS Domain Chemical Probe.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
7L3V
 
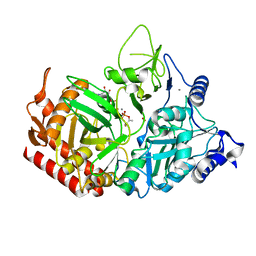 | | PEPCK MMQX structure 120ms post-mixing with oxaloacetic acid | | Descriptor: | CARBON DIOXIDE, GUANOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Clinger, J.A, Moreau, D.W, McLeod, M.J, Holyoak, T, Thorne, R.E. | | Deposit date: | 2020-12-18 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Millisecond mix-and-quench crystallography (MMQX) enables time-resolved studies of PEPCK with remote data collection.
Iucrj, 8, 2021
|
|
2O38
 
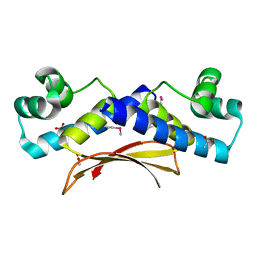 | | Putative XRE Family Transcriptional Regulator | | Descriptor: | ACETIC ACID, Hypothetical protein | | Authors: | Kim, Y, Joachimiak, A, Evdokimova, E, Kagan, O, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-11-30 | | Release date: | 2007-01-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The Crystal Structure of Putative XRE Family Transcriptional Regulator
To be Published
|
|
2UXA
 
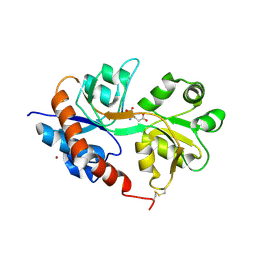 | | Crystal structure of the GluR2-flip ligand binding domain, r/g unedited. | | Descriptor: | GLUTAMATE RECEPTOR SUBUNIT GLUR2-FLIP, GLUTAMIC ACID, ZINC ION | | Authors: | Greger, I.H, Akamine, P, Khatri, L, Ziff, E.B. | | Deposit date: | 2007-03-27 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Developmentally Regulated, Combinatorial RNA Processing Modulates Ampa Receptor Biogenesis.
Neuron, 51, 2006
|
|
2O4H
 
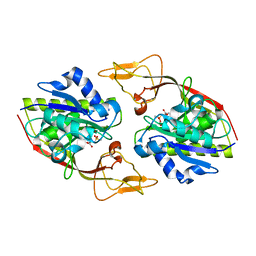 | | Human brain aspartoacylase complex with intermediate analog (N-phosphonomethyl-L-aspartate) | | Descriptor: | Aspartoacylase, N-[HYDROXY(METHYL)PHOSPHORYL]-L-ASPARTIC ACID, ZINC ION | | Authors: | Le Coq, J, Pavlovsky, A, Sanishvili, R, Viola, R.E. | | Deposit date: | 2006-12-04 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Examination of the Mechanism of Human Brain Aspartoacylase through the Binding of an Intermediate Analogue.
Biochemistry, 47, 2008
|
|
1CP5
 
 | | RECOMBINANT SPERM WHALE MYOGLOBIN L104F MUTANT (MET) | | Descriptor: | PROTEIN (MYOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Liong, E.C, Phillips Jr, G.N. | | Deposit date: | 1999-06-07 | | Release date: | 1999-06-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Waterproofing the heme pocket. Role of proximal amino acid side chains in preventing hemin loss from myoglobin.
J.Biol.Chem., 276, 2001
|
|
5E5Y
 
 | | Quasi-racemic snakin-1 in P1 before radiation damage | | Descriptor: | 1,2-ETHANEDIOL, D- snakin-1, FORMIC ACID, ... | | Authors: | Yeung, H, Squire, C.J, Yosaatmadja, Y, Panjikar, S, Baker, E.N, Harris, P.W.R, Brimble, M.A. | | Deposit date: | 2015-10-09 | | Release date: | 2016-05-18 | | Last modified: | 2016-07-20 | | Method: | X-RAY DIFFRACTION (1.506 Å) | | Cite: | Radiation Damage and Racemic Protein Crystallography Reveal the Unique Structure of the GASA/Snakin Protein Superfamily.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
2IFU
 
 | | Crystal Structure of a Gamma-SNAP from Danio rerio | | Descriptor: | SULFATE ION, gamma-snap | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Mccoy, J.G, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-09-21 | | Release date: | 2006-10-10 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and dynamics of gamma-SNAP: insight into flexibility of proteins from the SNAP family.
Proteins, 70, 2008
|
|
1O91
 
 | | Crystal Structure of a Collagen VIII NC1 Domain Trimer | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, COLLAGEN ALPHA 1(VIII) CHAIN, SULFATE ION | | Authors: | Kvansakul, M, Bogin, O, Hohenester, E, Yayon, A. | | Deposit date: | 2002-12-10 | | Release date: | 2003-11-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Collagen Alpha1(Viii) Nc1 Trimer.
Matrix Biol., 22, 2003
|
|
1OF1
 
 | | KINETICS AND CRYSTAL STRUCTURE OF THE HERPES SIMPLEX VIRUS TYPE 1 THYMIDINE KINASE INTERACTING WITH (SOUTH)-METHANOCARBA-THYMIDINE | | Descriptor: | (SOUTH)-METHANOCARBA-THYMIDINE, SULFATE ION, THYMIDINE KINASE | | Authors: | Claus, M.T, Schelling, P, Folkers, G, Marquez, V.E, Scapozza, L, Schulz, G.E. | | Deposit date: | 2003-04-03 | | Release date: | 2004-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biochemical and Structural Characterization of (South)-Methanocarbathymidine that Specifically Inhibits Growth of Herpes Simplex Virus Type 1 Thymidine Kinase-Transduced Osteosarcoma Cells
J.Biol.Chem., 279, 2004
|
|
6AF2
 
 | | Crystal structure of N-terminus deletion mutant of Mycobacterium avium diadenosine 5',5'''-P1,P4-tetraphosphate phosphorylase | | Descriptor: | DI(HYDROXYETHYL)ETHER, HIT domain-containing protein, PENTAETHYLENE GLYCOL, ... | | Authors: | Mori, S, Honda, N, Kim, H, Rimbara, E, Shibayama, K. | | Deposit date: | 2018-08-08 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Crystal structure of N-terminus deletion mutant of Mycobacterium avium diadenosine tetraphosphate phosphorylase
To Be Published
|
|
