8SF1
 
 | | Carbonic anhydrase II XFEL radiation damage RT | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Combs, J.E, Mckenna, R. | | Deposit date: | 2023-04-10 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | XFEL structure of carbonic anhydrase II: a comparative study of XFEL, NMR, X-ray and neutron structures.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
1BQU
 
 | | CYTOKYNE-BINDING REGION OF GP130 | | Descriptor: | GLYCEROL, PROTEIN (GP130), SULFATE ION | | Authors: | Bravo, J, Staunton, D, Heath, J.K, Jones, E.Y. | | Deposit date: | 1998-08-18 | | Release date: | 1998-08-26 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a cytokine-binding region of gp130.
EMBO J., 17, 1998
|
|
3MAO
 
 | | Crystal Structure of Human Methionine-R-Sulfoxide Reductase B1 (MsrB1) | | Descriptor: | FE (III) ION, MALONATE ION, Methionine-R-sulfoxide reductase B1, ... | | Authors: | Chaikuad, A, Shafqat, N, Yue, W.W, Savitsky, P, Krojer, T, Ugochukwu, E, Muniz, J.R.C, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-24 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal Structure of Human Methionine-R-Sulfoxide Reductase B1 (MsrB1)
To be Published
|
|
1JX9
 
 | | Penicillin Acylase, mutant | | Descriptor: | CALCIUM ION, penicillin G acylase alpha subunit, penicillin G acylase beta subunit | | Authors: | Hensgens, C.M.H, Keizer, E, Snijder, H.J, Dijkstra, B.W. | | Deposit date: | 2001-09-06 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural and kinetic studies on ligand binding in wild-type and active-site mutants of penicillin acylase.
Protein Eng.Des.Sel., 17, 2004
|
|
4PEI
 
 | | Dbr1 in complex with synthetic branched RNA analog | | Descriptor: | GLYCEROL, NICKEL (II) ION, RNA (5'-R(*(G46)P*U)-3'), ... | | Authors: | Montemayor, E.J, Katolik, A, Clark, N.E, Taylor, A.B, Schuermann, J.P, Combs, D.J, Johnsson, R, Holloway, S.P, Stevens, S.W, Damha, M.J, Hart, P.J. | | Deposit date: | 2014-04-23 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of lariat RNA recognition by the intron debranching enzyme Dbr1.
Nucleic Acids Res., 42, 2014
|
|
2UY8
 
 | | R92A mutant of Bacillus subtilis Oxalate Decarboxylase OxdC | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, OXALATE DECARBOXYLASE OXDC | | Authors: | Just, V.J, Burrell, M.R, Bowater, L, McRobbie, I, Stevenson, C.E.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2007-04-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Identity of the Active Site of Oxalate Decarboxylase and the Importance of the Stability of Active-Site Lid Conformations.
Biochem.J., 407, 2007
|
|
5DCE
 
 | |
4AK2
 
 | | Structure of BT4661, a SusE-like surface located polysaccharide binding protein from the Bacteroides thetaiotaomicron heparin utilisation locus | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, BT_4661, SODIUM ION | | Authors: | Lowe, E.C, Basle, A, Czjzek, M, Thomas, S, Murray, H, Firbank, S.J, Bolam, D.N. | | Deposit date: | 2012-02-21 | | Release date: | 2013-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | How members of the human gut microbiota overcome the sulfation problem posed by glycosaminoglycans.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3MBY
 
 | | Ternary complex of DNA Polymerase BETA with template base A and 8oxodGTP in the active site with a dideoxy terminated primer | | Descriptor: | 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Batra, V.K, Beard, W.A, Hou, E.W, Pedersen, L.C, Prasad, R, Wilson, S.H. | | Deposit date: | 2010-03-26 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutagenic conformation of 8-oxo-7,8-dihydro-2'-dGTP in the confines of a DNA polymerase active site.
Nat.Struct.Mol.Biol., 17, 2010
|
|
8SH4
 
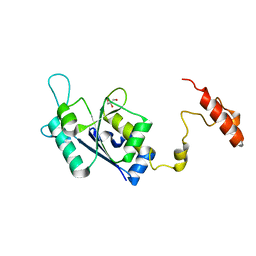 | | Crystal structure of the tRNA (m1G37) methyltransferase apoenzyme from Anaplasma phagocytophilum | | Descriptor: | GLYCEROL, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Jannotta, C, Edele, D, Levanti, D, Carson, M, Prucha, G, Caesar, J, Picchiello, C, Collins, K, Garland, E, Handley-Pendleton, J, Hernandez, V, Leffler, S, Williams, D, Stojanoff, V, Perez, A, Halloran, J, Bolen, R. | | Deposit date: | 2023-04-13 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the m1G37 tRNA methyltransferase apoenzyme from Anaplasma phagocytophilum
To Be Published
|
|
4AE1
 
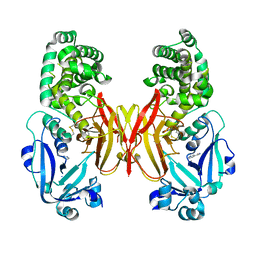 | |
4YNC
 
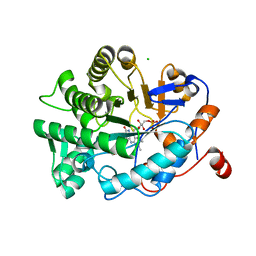 | | OYE1 W116A COMPLEXED WITH (Z)-METHYL-3-CYANO-3-PHENYLACRYLATE IN A NON PRODUCTIVE BINDING MODE | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, NADPH dehydrogenase 1, ... | | Authors: | Santangelo, S, Brenna, E, Stewart, J.D, Powell III, R.W. | | Deposit date: | 2015-03-09 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Opposite Enantioselectivity in the Bioreduction of (Z)-beta-Aryl-beta-cyanoacrylates Mediated by the Tryptophan 116 Mutants of Old Yellow Enzyme 1: Synthetic Approach to (R)- and (S)-beta-Aryl-gamma-lactams
Adv.Synth.Catal., 357, 2015
|
|
3D47
 
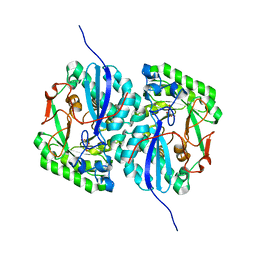 | | Crystal structure of L-rhamnonate dehydratase from Salmonella typhimurium complexed with Mg and D-malate | | Descriptor: | D-MALATE, MAGNESIUM ION, Putative galactonate dehydratase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-05-14 | | Release date: | 2008-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of L-rhamnonate dehydratase from Salmonella typhimurium complexed with Mg and D-malate.
To be Published
|
|
1JDO
 
 | |
3MG5
 
 | | Core-streptavidin mutant F130L in complex with biotin | | Descriptor: | BIOTIN, GLYCEROL, Streptavidin | | Authors: | Le Trong, I, Baugh, L, Stayton, P.S, Lybrand, T.P, Stenkamp, R.E. | | Deposit date: | 2010-04-05 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A distal point mutation in the streptavidin-biotin complex preserves structure but diminishes binding affinity: experimental evidence of electronic polarization effects?
Biochemistry, 49, 2010
|
|
4AJ9
 
 | | Catalase 3 from Neurospora crassa | | Descriptor: | ACETATE ION, CATALASE-3, PENTAETHYLENE GLYCOL, ... | | Authors: | Zarate-Romero, A, Rudino-Pinera, E. | | Deposit date: | 2012-02-16 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray driven reduction of Cpd I of Catalase-3 from N. crassa reveals differential sensitivity of active sites and formation of ferrous state.
Arch.Biochem.Biophys., 666, 2019
|
|
2UYV
 
 | |
2IF7
 
 | | Crystal Structure of NTB-A | | Descriptor: | CALCIUM ION, CHLORIDE ION, SLAM family member 6 | | Authors: | Cao, E, Ramagopal, U.A, Fedorov, A.A, Fedorov, E.V, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2006-09-20 | | Release date: | 2006-10-17 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | NTB-A Receptor Crystal Structure: Insights into Homophilic Interactions in the Signaling Lymphocytic Activation Molecule Receptor Family.
Immunity, 25, 2006
|
|
4DR1
 
 | | Crystal structure of the apo 30S ribosomal subunit from Thermus thermophilus (HB8) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-16 | | Release date: | 2012-11-14 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | A structural basis for streptomycin-induced misreading of the genetic code.
Nat Commun, 4, 2013
|
|
1OGA
 
 | | A structural basis for immunodominant human T-cell receptor recognition. | | Descriptor: | BETA-2-MICROGLOBULIN, GILGFVFTL, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Stewart-Jones, G.B.E, McMichael, A.J, Bell, J.I, Stuart, D.I, Jones, E.Y. | | Deposit date: | 2003-04-28 | | Release date: | 2003-06-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Structural Basis for Immunodominant Human T Cell Receptor Recognition
Nat.Immunol., 4, 2003
|
|
1UGH
 
 | | CRYSTAL STRUCTURE OF HUMAN URACIL-DNA GLYCOSYLASE IN COMPLEX WITH A PROTEIN INHIBITOR: PROTEIN MIMICRY OF DNA | | Descriptor: | PROTEIN (URACIL-DNA GLYCOSYLASE INHIBITOR), PROTEIN (URACIL-DNA GLYCOSYLASE) | | Authors: | Mol, C.D, Arvai, A.S, Sanderson, R.J, Slupphaug, G, Kavli, B, Krokan, H.E, Mosbaugh, D.W, Tainer, J.A. | | Deposit date: | 1999-02-05 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human uracil-DNA glycosylase in complex with a protein inhibitor: protein mimicry of DNA.
Cell(Cambridge,Mass.), 82, 1995
|
|
3MEL
 
 | | Crystal Structure of Thiamin pyrophosphokinase family protein from Enterococcus faecalis, Northeast Structural Genomics Consortium Target EfR150 | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, THIAMINE DIPHOSPHATE, ... | | Authors: | Kuzin, A, Abasidze, M, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Foote, E.L, Maglaqui, M, Zhao, L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.788 Å) | | Cite: | Northeast Structural Genomics Consortium Target EfR150
To be Published
|
|
2IHY
 
 | | Structure of the Staphylococcus aureus putative ATPase subunit of an ATP-binding cassette (ABC) transporter | | Descriptor: | ABC transporter, ATP-binding protein, SULFATE ION | | Authors: | McGrath, T.E, Yu, C.S, Romanov, V, Lam, R, Dharamsi, A, Virag, C, Mansoury, K, Thambipillai, D, Richards, D, Guthrie, J, Edwards, A.M, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2006-09-27 | | Release date: | 2007-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the Staphylococcus aureus putative ATPase subunit of an ATP-binding cassette (ABC) transporter
To be Published
|
|
8RU5
 
 | | ATPase family AAA domain containing 2 with crystallization epitope mutations V1022R:Q1027E | | Descriptor: | 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2 | | Authors: | Fairhead, M, Strain-Damerell, C, Ye, M, Mackinnon, S.R, Pinkas, D, MacLean, E.M, Koekemoer, L, Damerell, D, Krojer, T, Arrowsmith, C.H, Edwards, A, Bountra, C, Yue, W, Burgess-Brown, N, Marsden, B, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-01-30 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | A fast, parallel method for efficiently exploring crystallization behaviour of large numbers of protein variants
To Be Published
|
|
3MFG
 
 | |
