5OEJ
 
 | | Structure of Tra1 subunit within the chromatin modifying complex SAGA | | Descriptor: | Tra1 subunit within the chromatin modifying complex SAGA | | Authors: | Sharov, G, Voltz, K, Durand, A, Kolesnikova, O, Papai, G, Myasnikov, A.G, Dejaegere, A, Ben-Shem, A, Schultz, P. | | Deposit date: | 2017-07-07 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structure of the transcription activator target Tra1 within the chromatin modifying complex SAGA.
Nat Commun, 8, 2017
|
|
2WPS
 
 | | Salmonella enterica SadA 483-523 fused to GCN4 adaptors (SadAK3b-V2, out-of-register fusion) | | Descriptor: | CHLORIDE ION, TRIMERIC AUTOTRANSPORTER ADHESIN FRAGMENT | | Authors: | Hartmann, M.D, Ridderbusch, O, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2009-08-09 | | Release date: | 2009-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Coiled-Coil Motif that Sequesters Ions to the Hydrophobic Core.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8JSH
 
 | |
1IJ3
 
 | |
3V4G
 
 | | 1.60 Angstrom resolution crystal structure of an arginine repressor from Vibrio vulnificus CMCP6 | | Descriptor: | Arginine repressor | | Authors: | Halavaty, A.S, Minasov, G, Filippova, E, Shuvalova, L, Winsor, J, Dubrovska, I, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-14 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1.60 Angstrom resolution crystal structure of an arginine repressor from Vibrio vulnificus CMCP6
To be Published
|
|
7FTU
 
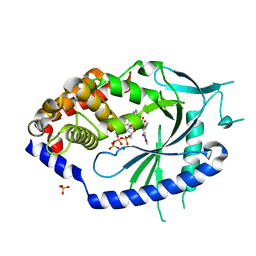 | | Crystal Structure of human cyclic GMP-AMP synthase in complex with (Z)-N-[4-(4-chlorophenyl)sulfonylphenyl]-2-cyano-3-hydroxy-3-(5-methyl-1,2-oxazol-4-yl)prop-2-enamide | | Descriptor: | (2Z)-N-[4-(4-chlorobenzene-1-sulfonyl)phenyl]-2-cyano-3-hydroxy-3-(5-methyl-1,2-oxazol-4-yl)prop-2-enamide, ADENOSINE-5'-TRIPHOSPHATE, Cyclic GMP-AMP synthase, ... | | Authors: | Leibrock, L, Benz, J, Groebke-Zbinden, K, Rudolph, M.G. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of a human cyclic GMP-AMP synthase complex
To be published
|
|
7FTX
 
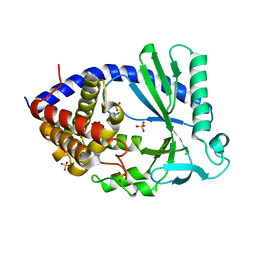 | |
8FDU
 
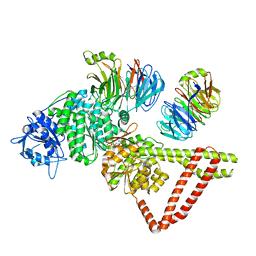 | | Engineered human dynein motor domain in the microtubule-unbound state with LIS1 complex in the buffer containing ATP-Vi (local refined on AAA3-AAA5 and LIS1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cytoplasmic dynein 1 heavy chain 1,Serine--tRNA ligase, Platelet-activating factor acetylhydrolase IB subunit beta,Platelet-activating factor acetylhydrolase IB subunit beta,human LIS1 protein with a SNAP tag | | Authors: | Ton, W, Wang, Y, Chai, P. | | Deposit date: | 2022-12-04 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Microtubule-binding-induced allostery triggers LIS1 dissociation from dynein prior to cargo transport.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3JBY
 
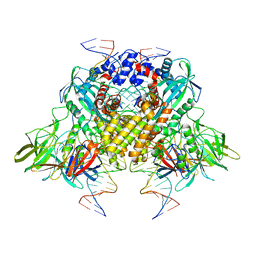 | | Cryo-electron microscopy structure of RAG Paired Complex (C2 symmetry) | | Descriptor: | '-D(P*GP*AP*TP*CP*TP*GP*GP*CP*CP*TP*GP*TP*CP*TP*TP*A)-3', 5'-D(P*CP*AP*CP*AP*GP*TP*GP*CP*TP*AP*CP*AP*GP*AP*C)-3', CALCIUM ION, ... | | Authors: | Ru, H, Chambers, M.G, Fu, T.-M, Tong, A.B, Liao, M, Wu, H. | | Deposit date: | 2015-10-22 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular Mechanism of V(D)J Recombination from Synaptic RAG1-RAG2 Complex Structures.
Cell(Cambridge,Mass.), 163, 2015
|
|
8FDT
 
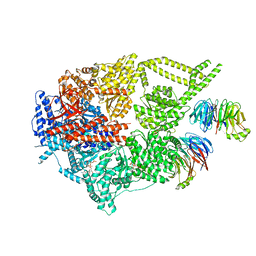 | | Engineered human dynein motor domain in the microtubule-unbound state with LIS1 complex in the buffer containing ATP-Vi | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cytoplasmic dynein 1 heavy chain 1,Serine--tRNA ligase, ... | | Authors: | Ton, W, Wang, Y, Chai, P. | | Deposit date: | 2022-12-04 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Microtubule-binding-induced allostery triggers LIS1 dissociation from dynein prior to cargo transport.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6HHQ
 
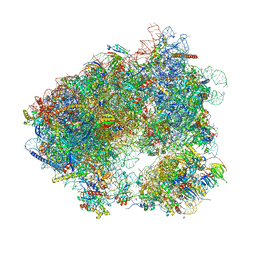 | | Crystal structure of compound C45 bound to the yeast 80S ribosome | | Descriptor: | (3~{R})-3-[(1~{S})-2-[(1~{S},4~{a}~{R},6~{S},7~{S},8~{a}~{R})-6,7-bis(chloranyl)-5,5,8~{a}-trimethyl-2-methylidene-3,4,4~{a},6,7,8-hexahydro-1~{H}-naphthalen-1-yl]-1-oxidanyl-ethyl]pyrrolidine-2,5-dione, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Pellegrino, S, Vanderwal, C.D, Yusupov, M. | | Deposit date: | 2018-08-28 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.10000038 Å) | | Cite: | Understanding the role of intermolecular interactions between lissoclimides and the eukaryotic ribosome.
Nucleic Acids Res., 47, 2019
|
|
3COJ
 
 | |
7NAS
 
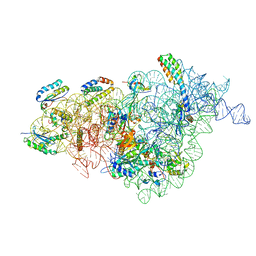 | | Bacterial 30S ribosomal subunit assembly complex state A (multibody refinement for body domain of 30S ribosome) | | Descriptor: | 16S rRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
4JX7
 
 | | Crystal structure of Pim1 kinase in complex with inhibitor 2-[(trans-4-aminocyclohexyl)amino]-4-{[3-(trifluoromethyl)phenyl]amino}pyrido[4,3-d]pyrimidin-5(6H)-one | | Descriptor: | 2-[(trans-4-aminocyclohexyl)amino]-4-{[3-(trifluoromethyl)phenyl]amino}pyrido[4,3-d]pyrimidin-5(6H)-one, PIM1 consensus peptide, Serine/threonine-protein kinase pim-1 | | Authors: | Lee, S.J, Han, B.G, Cho, J.W, Choi, J.S, Lee, J.K, Song, H.J, Koh, J.S, Lee, B.I. | | Deposit date: | 2013-03-27 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of pim1 kinase in complex with a pyrido[4,3-d]pyrimidine derivative suggests a unique binding mode.
Plos One, 8, 2013
|
|
1J8I
 
 | | Solution Structure of Human Lymphotactin | | Descriptor: | Lymphotactin | | Authors: | Kuloglu, E.S, McCaslin, D.R, Markley, J.L, Pauza, C.D, Volkman, B.F. | | Deposit date: | 2001-05-21 | | Release date: | 2001-10-24 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Monomeric solution structure of the prototypical 'C' chemokine lymphotactin.
Biochemistry, 40, 2001
|
|
5Q1B
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 4-{[(2S)-2-cyclohexyl-2-{5,6-difluoro-2-[4-(1,3-thiazol-2-yl)phenyl]-1H-benzimidazol-1-yl}acetyl]amino}benzoic acid, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5NTW
 
 | |
4KMH
 
 | |
5NU1
 
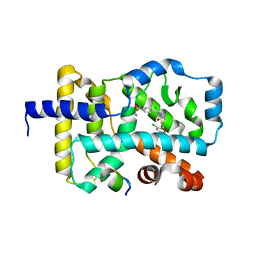 | |
6W7W
 
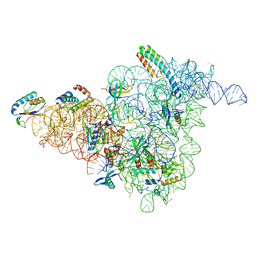 | | 30S-Inactive-low-Mg2+ Class B | | Descriptor: | 16S rRNA, 30S ribosomal protein S12, 30S ribosomal protein S15, ... | | Authors: | Jahagirdar, D, Jha, V, Basu, B, Gomez-Blanco, J, Vargas, J, Ortega, J. | | Deposit date: | 2020-03-19 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Alternative conformations and motions adopted by 30S ribosomal subunits visualized by cryo-electron microscopy.
Rna, 26, 2020
|
|
5Q1E
 
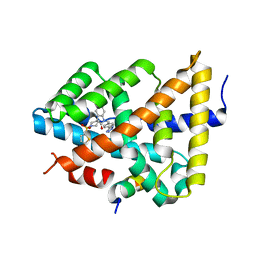 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 5-bromo-1-{[4-(1H-tetrazol-5-yl)phenyl]methyl}-1'-(thiophene-2-sulfonyl)spiro[indole-3,4'-piperidin]-2(1H)-one, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0L
 
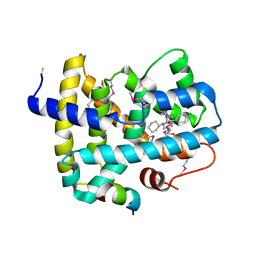 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-N,2-dicyclohexyl-2-{2-[4-(hydroxymethyl)phenyl]-1H-benzimidazol-1-yl}acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0Y
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-N,2-dicyclohexyl-2-{5,6-difluoro-2-[6-(1H-pyrazol-1-yl)pyridin-3-yl]-1H-benzimidazol-1-yl}acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q1F
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, trans-4-({(2S)-2-cyclohexyl-2-[2-(2,6-dimethoxypyridin-3-yl)-5-fluoro-1H-benzimidazol-1-yl]acetyl}amino)cyclohexane-1-carboxylic acid | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
2XZR
 
 | | Escherichia coli Immunoglobulin-binding protein EibD 391-438 FUSED TO GCN4 ADAPTORS | | Descriptor: | CHLORIDE ION, IMMUNOGLOBULIN-BINDING PROTEIN EIBD | | Authors: | Hartmann, M.D, Hernandez Alvarez, B, Ridderbusch, O, Deiss, S, Lupas, A.N. | | Deposit date: | 2010-11-28 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure of E. Coli Igg-Binding Protein D Suggests a General Model for Bending and Binding in Trimeric Autotransporter Adhesins.
Structure, 19, 2011
|
|
