1LS4
 
 | |
1I5J
 
 | |
1C55
 
 | |
5H7P
 
 | | NMR structure of the Vta1NTD-Did2(176-204) complex | | Descriptor: | Vacuolar protein sorting-associated protein VTA1, Vacuolar protein-sorting-associated protein 46 | | Authors: | Shen, J, Yang, Z, Wild, C.J. | | Deposit date: | 2016-11-20 | | Release date: | 2016-12-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR studies on the interactions between yeast Vta1 and Did2 during the multivesicular bodies sorting pathway
Sci Rep, 6, 2016
|
|
5KGQ
 
 | | NMR structure and dynamics of Q4DY78, a conserved kinetoplasid-specific protein from Trypanosoma cruzi | | Descriptor: | Uncharacterized protein | | Authors: | D'Andrea, E.D, Retel, J.S, Diehl, A, Schmieder, P, Oschkinat, H, Pires, J.R. | | Deposit date: | 2016-06-13 | | Release date: | 2017-07-05 | | Last modified: | 2024-06-12 | | Method: | SOLUTION NMR | | Cite: | NMR structure and dynamics of Q4DY78, a conserved kinetoplasid-specific protein from Trypanosoma cruzi.
J.Struct.Biol., 213, 2021
|
|
6JCE
 
 | | NMR solution and X-ray crystal structures of a DNA containing both right-and left-handed parallel-stranded G-quadruplexes | | Descriptor: | 29-mer DNA | | Authors: | Winnerdy, F.R, Bakalar, B, Maity, A, Vandana, J.J, Mechulam, Y, Schmitt, E, Phan, A.T. | | Deposit date: | 2019-01-28 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution and X-ray crystal structures of a DNA molecule containing both right- and left-handed parallel-stranded G-quadruplexes.
Nucleic Acids Res., 47, 2019
|
|
1N1K
 
 | | NMR Structure for d(CCGCGG)2 | | Descriptor: | 5'-D(P*CP*CP*GP*CP*GP*G)-3' | | Authors: | Monleon, D, Celda, B. | | Deposit date: | 2002-10-18 | | Release date: | 2002-10-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR study of hexanucleotide d(CCGCGG)(2) containing two triplet repeats of
fragile X syndrome.
Biochem.Biophys.Res.Commun., 303, 2003
|
|
5XJK
 
 | |
1MOT
 
 | | NMR Structure Of Extended Second Transmembrane Domain Of Glycine Receptor alpha1 Subunit in SDS Micelles | | Descriptor: | Glycine Receptor alpha-1 CHAIN | | Authors: | Yushmanov, V.E, Mandal, P.K, Liu, Z, Tang, P, Xu, Y. | | Deposit date: | 2002-09-09 | | Release date: | 2003-09-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure and Backbone Dynamics of the Extended Second Transmembrane Domain of the Human Neuronal Glycine Receptor Alpha1 Subunit
Biochemistry, 42, 2003
|
|
1L1K
 
 | |
1NJQ
 
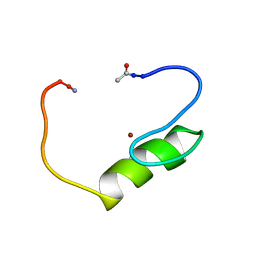 | | NMR structure of the single QALGGH zinc finger domain from Arabidopsis thaliana SUPERMAN protein | | Descriptor: | ZINC ION, superman protein | | Authors: | Isernia, C, Bucci, E, Leone, M, Zaccaro, L, Di Lello, P, Digilio, G, Esposito, S, Saviano, M, Di Blasio, B, Pedone, C, Pedone, P.V, Fattorusso, R. | | Deposit date: | 2003-01-02 | | Release date: | 2003-03-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Single QALGGH Zinc Finger Domain from the Arabidopsis thaliana SUPERMAN Protein.
Chembiochem, 4, 2003
|
|
1Q2I
 
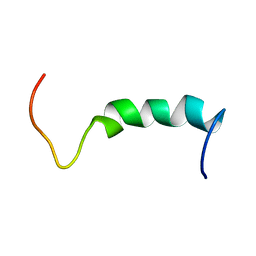 | | NMR SOLUTION STRUCTURE OF A PEPTIDE FROM THE MDM-2 BINDING DOMAIN OF THE P53 PROTEIN THAT IS SELECTIVELY CYTOTOXIC TO CANCER CELLS | | Descriptor: | PNC27 | | Authors: | Rosal, R, Pincus, M.R, Brandt-Rauf, P.W, Fine, R.L, Wang, H. | | Deposit date: | 2003-07-24 | | Release date: | 2004-03-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a peptide from the mdm-2 binding domain of the p53 protein that is selectively cytotoxic to cancer cells
Biochemistry, 43, 2004
|
|
1HLL
 
 | |
1Q2F
 
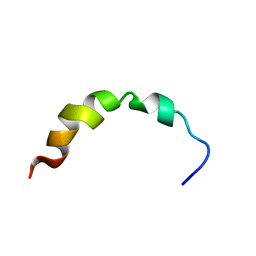 | | NMR SOLUTION STRUCTURE OF A PEPTIDE FROM THE MDM-2 BINDING DOMAIN OF THE P53 PROTEIN THAT IS SELECTIVELY CYTOTOXIC TO CANCER CELLS | | Descriptor: | PNC27 | | Authors: | Rosal, R, Pincus, M.R, Brandt-Rauf, P.W, Fine, R.L, Wang, H. | | Deposit date: | 2003-07-24 | | Release date: | 2004-03-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a peptide from the mdm-2 binding domain of the p53 protein that is selectively cytotoxic to cancer cells
Biochemistry, 43, 2004
|
|
1RJT
 
 | |
1S9L
 
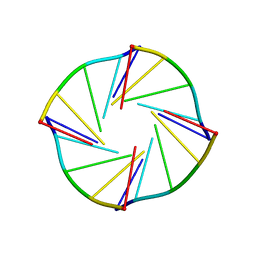 | | NMR Solution Structure of a Parallel LNA Quadruplex | | Descriptor: | 5'-((TLN)P*(LCG)P*(LCG)P*(LCG)P*(TLN))-3' | | Authors: | Randazzo, A, Esposito, V, Ohlenschlager, O, Ramachandran, R, Mayola, L. | | Deposit date: | 2004-02-05 | | Release date: | 2004-06-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a parallel LNA quadruplex.
Nucleic Acids Res., 32, 2004
|
|
1T23
 
 | | NMR Solution Structure of the Archaebacterial Chromosomal Protein MC1 | | Descriptor: | Chromosomal protein MC1 | | Authors: | Paquet, F, Culard, F, Barbault, F, Maurizot, J.C, Lancelot, G. | | Deposit date: | 2004-04-20 | | Release date: | 2004-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of the Archaebacterial Chromosomal Protein MC1 Reveals a New Protein Fold
Biochemistry, 43, 2004
|
|
1HOD
 
 | |
1HOF
 
 | |
1PG9
 
 | | NMR Solution Structure of an Oxaliplatin 1,2-d(GG) Intrastrand Cross-Link in a DNA Dodecamer Duplex | | Descriptor: | 5'-D(*CP*CP*TP*CP*AP*GP*GP*CP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*GP*CP*CP*TP*GP*AP*GP*G)-3', CYCLOHEXANE-1(R),2(R)-DIAMINE-PLATINUM(II) | | Authors: | Wu, Y, Pradhan, P, Havener, J, Chaney, S.G, Campbel, S.L. | | Deposit date: | 2003-05-28 | | Release date: | 2004-07-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of an oxaliplatin 1,2-d(GG) intrastrand cross-link in a DNA dodecamer duplex
J.Mol.Biol., 341, 2004
|
|
1PGC
 
 | | NMR Solution Structure of an Oxaliplatin 1,2-d(GG) Intrastrand Cross-Link in a DNA Dodecamer Duplex | | Descriptor: | 5'-D(*CP*CP*TP*CP*AP*GP*GP*CP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*GP*CP*CP*TP*GP*AP*GP*G)-3', CYCLOHEXANE-1(R),2(R)-DIAMINE-PLATINUM(II) | | Authors: | Wu, Y, Pradhan, P, Havener, J, Chaney, S.G, Campbel, S.L. | | Deposit date: | 2003-05-28 | | Release date: | 2004-07-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of an oxaliplatin 1,2-d(GG) intrastrand cross-link in a DNA dodecamer duplex
J.Mol.Biol., 341, 2004
|
|
1F5Y
 
 | | NMR STRUCTURE OF A CONCATEMER OF THE FIRST AND SECOND LIGAND-BINDING MODULES OF THE HUMAN LDL RECEPTOR | | Descriptor: | CALCIUM ION, LOW-DENSITY LIPOPROTEIN RECEPTOR | | Authors: | Kurniawan, N.D, Atkins, A.R, Brereton, I.M, Kroon, P.A, Smith, R. | | Deposit date: | 2000-06-18 | | Release date: | 2000-08-30 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a concatemer of the first and second ligand-binding modules of the human low-density lipoprotein receptor.
Protein Sci., 9, 2000
|
|
1NAU
 
 | | NMR Solution Structure of the Glucagon Antagonist [desHis1, desPhe6, Glu9]Glucagon Amide in the Presence of Perdeuterated Dodecylphosphocholine Micelles | | Descriptor: | Glucagon | | Authors: | Ying, J, Ahn, J.-M, Jacobsen, N.E, Brown, M.F, Hruby, V.J. | | Deposit date: | 2002-11-28 | | Release date: | 2003-03-18 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of the Glucagon Antagonist [desHis1, desPhe6, Glu9]Glucagon Amide in the Presence of Perdeuterated Dodecylphosphocholine Micelles
Biochemistry, 42, 2003
|
|
1PQQ
 
 | | NMR Structure of a Cyclic Polyamide-DNA Complex | | Descriptor: | 45-(3-AMINOPROPYL)-5,11,22,28,34-PENTAMETHYL-3,9,15,20,26,32,38,43-OCTAOXO-2,5,8,14,19,22,25,28,31,34,37,42,45,48-TETRADECAAZA-11-AZONIAHEPTACYCLO[42.2.1.1~4,7~.1~10,13~.1~21,24~.1~27,30~.1~33,36~]DOPENTACONTA-1(46),4(52),6,10(51),12,21(50),23,27(49),29,33(48),35,44(47)-DODECAENE, 5'-D(*CP*GP*CP*TP*AP*AP*CP*AP*GP*GP*C)-3', 5'-D(*GP*CP*CP*TP*GP*TP*TP*AP*GP*CP*G)-3' | | Authors: | Zhang, Q, Dwyer, T.J, Tsui, V, Case, D.A, Cho, J, Dervan, P.B, Wemmer, D.E. | | Deposit date: | 2003-06-18 | | Release date: | 2004-06-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of a Cyclic Polyamide-DNA Complex.
J.Am.Chem.Soc., 126, 2004
|
|
1PN5
 
 | | NMR structure of the NALP1 Pyrin domain (PYD) | | Descriptor: | NACHT-, LRR- and PYD-containing protein 2 | | Authors: | Hiller, S, Kohl, A, Fiorito, F, Herrmann, T, Wider, G, Tschopp, J, Grutter, M.G, Wuthrich, K. | | Deposit date: | 2003-06-12 | | Release date: | 2003-10-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the apoptosis- and inflammation-related NALP1 pyrin domain
Structure, 11, 2003
|
|
