4ASH
 
 | |
3UQS
 
 | |
3SFG
 
 | |
4X2V
 
 | |
4NRU
 
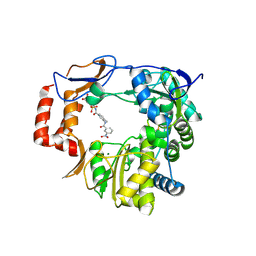 | | Murine Norovirus RNA-dependent-RNA-polymerase in complex with Compound 6, a suramin derivative | | Descriptor: | 4-({4-methyl-3-[(3-nitrobenzoyl)amino]benzoyl}amino)naphthalene-1,5-disulfonic acid, MAGNESIUM ION, RNA dependent RNA polymerase | | Authors: | Milani, M, Croci, R, Pezzullo, M, Tarantino, D, Mastrangelo, E, Bolognesi, M. | | Deposit date: | 2013-11-27 | | Release date: | 2014-10-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural bases of norovirus RNA dependent RNA polymerase inhibition by novel suramin-related compounds.
Plos One, 9, 2014
|
|
8A8X
 
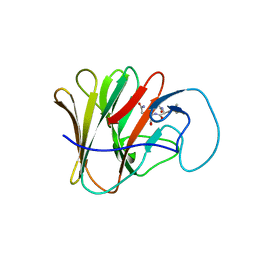 | | TRIM7 PRYSPRY in complex with a MNV1-NS3 peptide HDDFGLQ | | Descriptor: | E3 ubiquitin-protein ligase TRIM7, MNV1-NS3 C term peptide | | Authors: | Luptak, J. | | Deposit date: | 2022-06-24 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | TRIM7 Restricts Coxsackievirus and Norovirus Infection by Detecting the C-Terminal Glutamine Generated by 3C Protease Processing.
Viruses, 14, 2022
|
|
4O4R
 
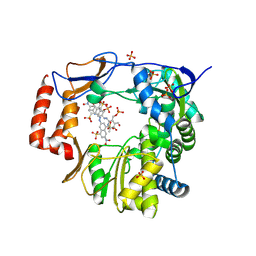 | | Murine Norovirus RdRp in complex with PPNDS | | Descriptor: | 3-[(E)-{4-formyl-5-hydroxy-6-methyl-3-[(phosphonooxy)methyl]pyridin-2-yl}diazenyl]-7-nitronaphthalene-1,5-disulfonic acid, RNA-dependent-RNA-polymerase, SULFATE ION | | Authors: | Croci, R, Tarantino, D, Milani, M, Pezzullo, M, Bolognesi, M, Mastrangelo, E. | | Deposit date: | 2013-12-19 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | PPNDS inhibits murine Norovirus RNA-dependent RNA-polymerase mimicking two RNA stacking bases.
Febs Lett., 588, 2014
|
|
4X2Y
 
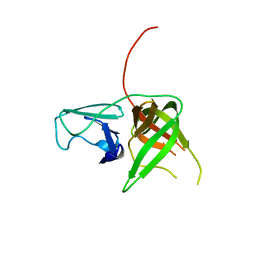 | | Crystal structure of a chimeric Murine Norovirus NS6 protease (inactive C139A mutant) in which the P4-P4 prime residues of the cleavage junction in the extended C-terminus have been replaced by the corresponding residues from the NS2-3 junction. | | Descriptor: | NS6 Protease,NS6 Protease | | Authors: | Leen, E.N, Pfeil, M.-P, Fernandes, H, Curry, S. | | Deposit date: | 2014-11-27 | | Release date: | 2015-02-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.417 Å) | | Cite: | Structure determination of Murine Norovirus NS6 proteases with C-terminal extensions designed to probe protease-substrate interactions.
Peerj, 3, 2015
|
|
3UR0
 
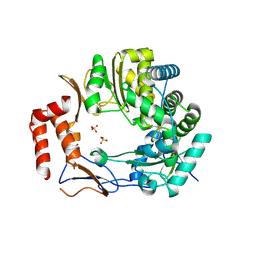 | | Crystal structures of murine norovirus RNA-dependent RNA polymerase in complex with Suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFONIC ACID, RNA-dependent RNA polymerase, SULFATE ION | | Authors: | Milani, M, Mastrangelo, E, Bolognesi, M. | | Deposit date: | 2011-11-21 | | Release date: | 2012-05-02 | | Last modified: | 2012-05-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-Based Inhibition of Norovirus RNA-Dependent RNA Polymerases.
J.Mol.Biol., 419, 2012
|
|
4X2X
 
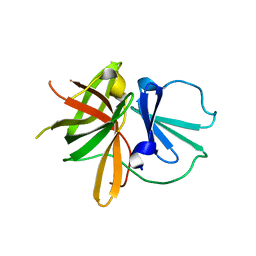 | |
3QID
 
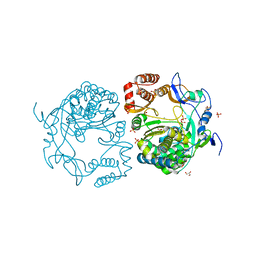 | | Crystal structures and functional analysis of murine norovirus RNA-dependent RNA polymerase | | Descriptor: | GLYCEROL, MANGANESE (III) ION, RNA dependent RNA polymerase, ... | | Authors: | Kim, K.H, Intekhab, A, Lee, J.H. | | Deposit date: | 2011-01-27 | | Release date: | 2011-12-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of murine norovirus-1 RNA-dependent RNA polymerase.
J.Gen.Virol., 92, 2011
|
|
3SFU
 
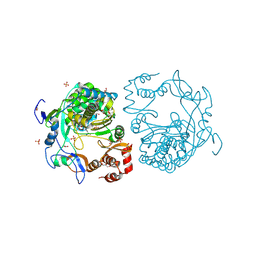 | | crystal structure of murine norovirus RNA dependent RNA polymerase in complex with ribavirin | | Descriptor: | 1-(beta-D-ribofuranosyl)-1H-1,2,4-triazole-3-carboxamide, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kim, K.H, Alam, I. | | Deposit date: | 2011-06-14 | | Release date: | 2012-05-09 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Crystal structures of murine norovirus-1 RNA-dependent RNA polymerase in complex with 2-thiouridine or ribavirin.
Virology, 426, 2012
|
|
3NAI
 
 | | Crystal structures and functional analysis of murine norovirus RNA-dependent RNA polymerase | | Descriptor: | 5-FLUOROURACIL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kim, K.H, Lee, J.H, Alam, I, Park, Y, Kang, S. | | Deposit date: | 2010-06-02 | | Release date: | 2011-06-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Crystal structures and functional analysis of murine norovirus RNA-dependent RNA polymerase
To be Published
|
|
3UPF
 
 | | Crystal structure of murine norovirus RNA-dependent RNA polymerase bound to NF023 | | Descriptor: | 8-({3-[({3-[(4,6,8-trisulfonaphthalen-1-yl)carbamoyl]phenyl}carbamoyl)amino]benzoyl}amino)naphthalene-1,3,5-trisulfonic acid, RNA-dependent RNA polymerase, SULFATE ION | | Authors: | Milani, M, Mastrangelo, E, Bolognesi, M. | | Deposit date: | 2011-11-18 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Inhibition of Norovirus RNA-Dependent RNA Polymerases.
J.Mol.Biol., 419, 2012
|
|
4X2W
 
 | |
3NAH
 
 | | Crystal structures and functional analysis of murine norovirus RNA-dependent RNA polymerase | | Descriptor: | RNA dependent RNA polymerase, SULFATE ION | | Authors: | Kim, K.H, Lee, J.H, Alam, I, Park, Y, Kang, S. | | Deposit date: | 2010-06-02 | | Release date: | 2011-06-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structures and functional analysis of murine norovirus RNA-dependent RNA polymerase
To be Published
|
|
8A5M
 
 | | TRIM7 PRYSPRY in complex with a MNV1-NS6 peptide LEALEFQ | | Descriptor: | E3 ubiquitin-protein ligase TRIM7, MNV1-NS6 peptide LEALEFQ, PHOSPHATE ION | | Authors: | Luptak, J. | | Deposit date: | 2022-06-15 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.918 Å) | | Cite: | TRIM7 Restricts Coxsackievirus and Norovirus Infection by Detecting the C-Terminal Glutamine Generated by 3C Protease Processing.
Viruses, 14, 2022
|
|
5Y3D
 
 | |
2MCH
 
 | |
2M4G
 
 | | Solution structure of the Core Domain (11-85) of the Murine Norovirus VPg protein | | Descriptor: | Murine Norovirus VPg protein | | Authors: | Leen, E.N, Kwok, R, Birtley, J.R, Prater, S.N, Simpson, P.J, Matthews, S, Marchant, J, Curry, S. | | Deposit date: | 2013-02-05 | | Release date: | 2013-03-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures of the Compact Helical Core Domains of Feline Calicivirus and Murine Norovirus VPg Proteins.
J.Virol., 87, 2013
|
|
2MCD
 
 | |
