9AR1
 
 | | Structure of Epimerase Mth373 bound to uridine-5'-diphosphate-xylopyranose | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Carbone, V, Schofield, L.R, Sutherland-Smith, A.J, Ronimus, R.S. | | Deposit date: | 2024-02-22 | | Release date: | 2025-01-29 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structurally characterizing epimerases from the thermophilic pseudomurein-containing methanogen Methanothermobacter thermautotrophicus delta H
To Be Published
|
|
7M13
 
 | | Crystal structure of CJ1428, a GDP-D-GLYCERO-L-GLUCO-HEPTOSE SYNTHASE from campylobacter jejuni in the presence of NADPH | | Descriptor: | 1,2-ETHANEDIOL, GDP-L-fucose synthase, MAGNESIUM ION, ... | | Authors: | Anderson, T.K, Thoden, J.B, Raushel, F.M, Holden, H.M. | | Deposit date: | 2021-03-12 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Biosynthesis of d- glycero -l- gluco -Heptose in the Capsular Polysaccharides of Campylobacter jejuni .
Biochemistry, 60, 2021
|
|
8ZAV
 
 | | alcohol dehydrogenases KpADH mutant - S9Y/F161K | | Descriptor: | 1,2-ETHANEDIOL, NAD-dependent epimerase/dehydratase domain-containing protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, L, Ni, Y, Xu, G.C. | | Deposit date: | 2024-04-25 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Engineering alcohol dehydrogenases KpADH for enhanced organic-solvent tolerance and its molecular mechanisms
To Be Published
|
|
5TQM
 
 | | Cinnamoyl-CoA Reductase 1 from Sorghum bicolor in complex with NADP+ | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Cinnamoyl-CoA Reductase, GLYCEROL, ... | | Authors: | Sattler, S.A, Kang, C.H. | | Deposit date: | 2016-10-24 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and Biochemical Characterization of Cinnamoyl-CoA Reductases.
Plant Physiol., 173, 2017
|
|
6MFH
 
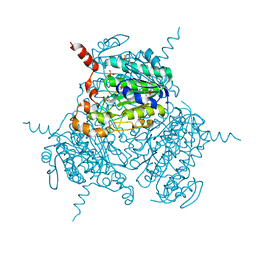 | | Mutated Uronate Dehydrogenase | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Mutated Uronate Dehydrogenase, PHOSPHATE ION | | Authors: | Sankaran, B, Pereira, J.H, Chan, V, Zwart, P.H, Wagschal, K. | | Deposit date: | 2018-09-11 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Chromohalobacter salixigens uronate dehydrogenase: Directed evolution for improved thermal stability and mutant CsUDH-inc X-ray crystal structure
Process Biochem, 2020
|
|
4LW8
 
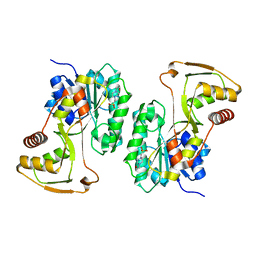 | |
2NNL
 
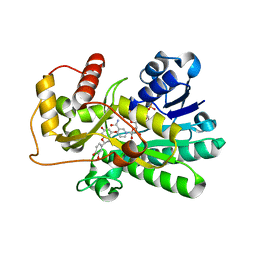 | | Binding of two substrate analogue molecules to dihydroflavonol-4-reductase alters the functional geometry of the catalytic site | | Descriptor: | (2S)-2-(3,4-DIHYDROXYPHENYL)-5,7-DIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, Dihydroflavonol 4-reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petit, P, Langlois D'Estaintot, B, Granier, T, Gallois, B. | | Deposit date: | 2006-10-24 | | Release date: | 2007-11-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding of two substrate analogue molecules to dihydroflavonol-4-reductase alters the functional geometry of the catalytic site
To be Published
|
|
6X3B
 
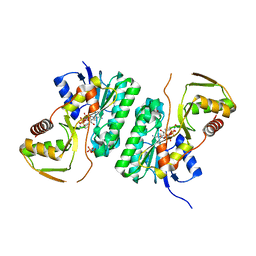 | |
1Z73
 
 | | Crystal Structure of E. coli ArnA dehydrogenase (decarboxylase) domain, S433A mutant | | Descriptor: | GLYCEROL, SULFATE ION, protein ArnA | | Authors: | Gatzeva-Topalova, P.Z, May, A.P, Sousa, M.C. | | Deposit date: | 2005-03-24 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Mechanism of ArnA: Conformational Change Implies Ordered Dehydrogenase Mechanism in Key Enzyme for Polymyxin Resistance
Structure, 13, 2005
|
|
4E5Y
 
 | |
4EJ0
 
 | |
7C3V
 
 | | Structure of a thermostable Alcohol dehydrogenase from Kluyveromyces polyspora(KpADH) | | Descriptor: | Alcohol dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Dai, W, Ni, Y, Xu, G, Liu, Y, Wang, Y, Zhou, J. | | Deposit date: | 2020-05-14 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.20042944 Å) | | Cite: | Structure of a thermostable Alcohol dehydrogenase from Kluyveromyces polyspora(KpADH)
To Be Published
|
|
2P5U
 
 | | Crystal structure of Thermus thermophilus HB8 UDP-glucose 4-epimerase complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase | | Authors: | Fu, Z.-Q, Chen, L, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhao, M, Dillard, B, Chrzas, J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of Thermus thermophilus HB8 UDP-glucose 4-epimerase complex with NAD
To be Published
|
|
4WOK
 
 | |
2P5Y
 
 | | Crystal structure of Thermus thermophilus HB8 UDP-glucose 4-epimerase complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase | | Authors: | Fu, Z.-Q, Chen, L, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhu, J, Swindell, J.T, Chrzas, J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of Thermus thermophilus HB8 UDP-glucose 4-epimerase complex with NAD
To be Published
|
|
8JQJ
 
 | | Crystal structure of carbonyl reductase SSCR mutant 1 from Sporobolomyces Salmonicolor | | Descriptor: | Aldehyde reductase 2 | | Authors: | Zhang, H.L, Li, Q, Liu, W.D, Chen, X, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2023-06-14 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Engineering a Carbonyl Reductase to Simultaneously Increase Activity Toward Bulky Ketone and Isopropanol for Dynamic Kinetic Asymmetric Reduction via Enzymatic Hydrogen Transfer
Acs Catalysis, 13, 2023
|
|
8JQK
 
 | | Crystal structure of a carbonyl reductase SSCR mutant from Sporobolomyces Salmonicolor | | Descriptor: | Aldehyde reductase 2 | | Authors: | Zhang, H.L, Li, Q, Liu, W.D, Chen, X, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2023-06-14 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Engineering a Carbonyl Reductase to Simultaneously Increase Activity Toward Bulky Ketone and Isopropanol for Dynamic Kinetic Asymmetric Reduction via Enzymatic Hydrogen Transfer
Acs Catalysis, 13, 2023
|
|
7CXT
 
 | |
8W3U
 
 | | Structure of Epimerase Mth373 from the thermophilic pseudomurein-containing methanogen Methanothermobacter thermautotrophicus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Carbone, V, Schofield, L.R, Sutherland-Smith, A.J, Ronimus, R.S. | | Deposit date: | 2024-02-22 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structurally characterizing epimerases from the thermophilic pseudomurein-containing methanogen Methanothermobacter thermautotrophicus delta H
To Be Published
|
|
8WXK
 
 | |
6ND7
 
 | | The crystal structure of TerB co-crystallized with polyporic acid | | Descriptor: | 2~3~,2~6~-dihydroxy[1~1~,2~1~:2~4~,3~1~-terphenyl]-2~2~,2~5~-dione, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Clinger, J.A, Elshahawi, S.I, Zhang, Y, Hall, R.P, Liu, Y, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2018-12-13 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structure and Function of Terfestatin Biosynthesis Enzymes TerB and TerC
To Be Published
|
|
6D2V
 
 | | Apo Structure of TerB, an NADP Dependent Oxidoreductase in the Terfestatin Biosynthesis Pathway | | Descriptor: | CHLORIDE ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, THIOCYANATE ION, ... | | Authors: | Clinger, J.A, Elshahawi, S.I, Zhang, Y, Hall, R.P, Liu, Y, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2018-04-14 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structure and Function of Terfestatin Biosynthesis Enzymes TerB and TerC
To Be Published
|
|
1A9Z
 
 | |
1A9Y
 
 | |
6NBR
 
 | |
