4W5U
 
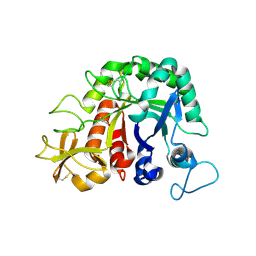 | |
5XEP
 
 | | Crystal structure of BRP39, a chitinase-like protein, at 2.6 Angstorm resolution | | Descriptor: | 1,2-ETHANEDIOL, Chitinase-3-like protein 1 | | Authors: | Mohanty, A.K, Fisher, A.J, Choudhary, S, Kaushik, J.K. | | Deposit date: | 2017-04-05 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of BRP39, a signalling glycoprotein expressed during mammary gland apoptosis.
To be published
|
|
5JH8
 
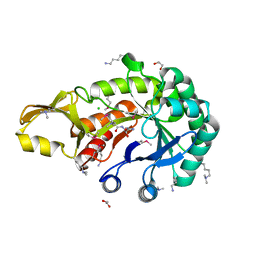 | | Crystal structure of chitinase from Chromobacterium violaceum ATCC 12472 | | Descriptor: | (2S)-2-(dimethylamino)-4-(methylselanyl)butanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Chang, C, Michalska, K, Tesar, C, Clancy, S, Joachimiak, A. | | Deposit date: | 2016-04-20 | | Release date: | 2016-05-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.018 Å) | | Cite: | Crystal structure of chitinase from Chromobacterium violaceum ATCC 12472
To Be Published
|
|
6VE1
 
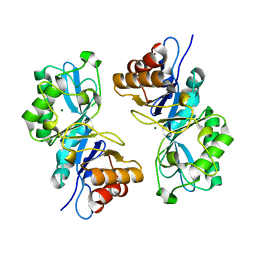 | |
2G41
 
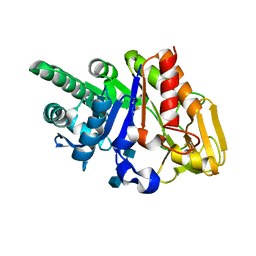 | | Crystal structure of the complex of sheep signalling glycoprotein with chitin trimer at 3.0A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SIGNAL PROCESSING PROTEIN, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Srivastava, D.B, Ethayathulla, A.S, Kumar, J, Bhushan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2006-02-21 | | Release date: | 2006-04-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Carbohydrate binding properties and carbohydrate induced conformational switch in sheep secretory glycoprotein (SPS-40): crystal structures of four complexes of SPS-40 with chitin-like oligosaccharides
J.Struct.Biol., 158, 2007
|
|
8C6Z
 
 | | necrotic enteritis associated Clostridium p. chitinase F8UNI4 in complex with inhibitor bisdionin C | | Descriptor: | 1,1'-PROPANE-1,3-DIYLBIS(3,7-DIMETHYL-3,7-DIHYDRO-1H-PURINE-2,6-DIONE), CADMIUM ION, COBALT (II) ION, ... | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Contribution and structural characterization of Chitinases to Clostridium perfringens Pathogenesis in Broilers
To Be Published
|
|
6EPB
 
 | | Structure of Chitinase 42 from Trichoderma harzianum | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Endochitinase 42, ... | | Authors: | Ramirez-Escudero, M, Jimenez-Ortega, E, Sanz-Aparicio, J. | | Deposit date: | 2017-10-11 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Use of chitin and chitosan to produce new chitooligosaccharides by chitinase Chit42: enzymatic activity and structural basis of protein specificity.
Microb. Cell Fact., 17, 2018
|
|
8DF1
 
 | | Chi3l1 bound by antibody C59 | | Descriptor: | C59 Fab Heavy chain, C59 Fab Light chain, Chitinase-3-like protein 1, ... | | Authors: | Wrapp, D, McLellan, J.S. | | Deposit date: | 2022-06-21 | | Release date: | 2023-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A novel humanized Chi3l1 blocking antibody attenuates acetaminophen-induced liver injury in mice.
Antib Ther, 6, 2023
|
|
5KZ6
 
 | | 1.25 Angstrom Crystal Structure of Chitinase from Bacillus anthracis. | | Descriptor: | CHLORIDE ION, Chitinase, SODIUM ION, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-22 | | Release date: | 2016-08-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.252 Å) | | Cite: | 1.25 Angstrom Crystal Structure of Chitinase from Bacillus anthracis.
To Be Published
|
|
7AKQ
 
 | |
6CAF
 
 | |
7Z64
 
 | | A GH18 from haloalkaliphilic bacterium unveils environment-dependent variations in the catalytic machinery of chitinases | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase, ... | | Authors: | Madhuprakash, J, Dalhus, B, Rohr, A.K, Bissaro, B, Vaaje-Kolstad, G, Sorlie, M, Eijsink, V.G. | | Deposit date: | 2022-03-10 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | A GH18 from haloalkaliphilic bacterium unveils environment-dependent variations in the catalytic machinery of chitinases
To Be Published
|
|
7Z65
 
 | | A GH18 from haloalkaliphilic bacterium unveils environment-dependent variations in the catalytic machinery of chitinases | | Descriptor: | 1,2-ETHANEDIOL, Chitinase, GH18 family | | Authors: | Madhuprakash, J, Dalhus, B, Rohr, A.K, Bissaro, B, Vaaje-Kolstad, G, Sorlie, M, Eijsink, V.G. | | Deposit date: | 2022-03-11 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.108 Å) | | Cite: | A GH18 from haloalkaliphilic bacterium unveils environment-dependent variations in the catalytic machinery of chitinases
To Be Published
|
|
3BXW
 
 | |
4MPK
 
 | | Crystal structure of the complex of buffalo signaling protein SPB-40 with N-acetylglucosamine at 2.65 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1, GLYCEROL | | Authors: | Yamini, S, Chaudhary, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-09-13 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the complex of buffalo signaling protein SPB-40 with N-acetylglucosamine at 2.65 A resolution
To be Published
|
|
7ZY9
 
 | |
7ZYA
 
 | | Structure of Chit33 from Trichoderma harzianum. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Endochitinase 33, ... | | Authors: | Jimenez-Ortega, E, Sanz-Aparicio, J. | | Deposit date: | 2022-05-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structure-Function Insights into the Fungal Endo -Chitinase Chit33 Depict its Mechanism on Chitinous Material.
Int J Mol Sci, 23, 2022
|
|
4N42
 
 | | Crystal structure of allergen protein scam1 from Scadoxus multiflorus | | Descriptor: | PHOSPHATE ION, Xylanase and alpha-amylase inhibitor protein isoform III | | Authors: | Singh, A, Kumar, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-10-08 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of allergen protein scam1 from Scadoxus multiflorus
To be published
|
|
3CHE
 
 | |
3IAN
 
 | | Crystal structure of a chitinase from Lactococcus lactis subsp. lactis | | Descriptor: | 1,2-ETHANEDIOL, Chitinase, SODIUM ION | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Miller, S, Ozyurt, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-14 | | Release date: | 2009-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a chitinase from Lactococcus lactis subsp. lactis
To be Published
|
|
6T9M
 
 | | Crystal structure of the Chitinase Domain of the Spore Coat Protein CotE from Clostridium difficile | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, Peptide in active site, ... | | Authors: | Whittingham, J.L, Dodson, E.J, Wilkinson, A.J. | | Deposit date: | 2019-10-28 | | Release date: | 2020-07-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of the GH18 domain of the bifunctional peroxiredoxin-chitinase CotE from Clostridium difficile.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6TSB
 
 | |
7XMH
 
 | | Crystal structure of a rice class IIIb chitinase, Oschib2 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Putative class III chitinase | | Authors: | Jun, T, Tomoya, T, Tomoyuki, N, Takayuki, O. | | Deposit date: | 2022-04-25 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Characterization of two rice GH18 chitinases belonging to family 8 of plant pathogenesis-related proteins.
Plant Sci., 326, 2023
|
|
3HU7
 
 | | Structural characterization and binding studies of a plant pathogenesis related protein heamanthin from haemanthus multiflorus reveal its dual inhibitory effects against xylanase and alpha-amylase | | Descriptor: | ACETATE ION, Haementhin, PHOSPHATE ION | | Authors: | Kumar, S, Singh, N, Sinha, M, Bhushan, A, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2009-06-13 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure determination and inhibition studies of a novel xylanase and alpha-amylase inhibitor protein (XAIP) from Scadoxus multiflorus.
Febs J., 277, 2010
|
|
8XBD
 
 | |
