3A9X
 
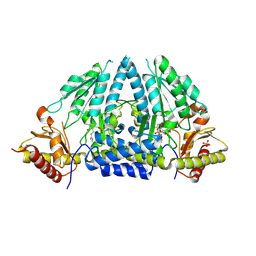 | | Crystal structure of rat selenocysteine lyase | | Descriptor: | PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, Selenocysteine lyase | | Authors: | Omi, R, Hirotsu, K. | | Deposit date: | 2009-11-08 | | Release date: | 2010-03-16 | | Last modified: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reaction mechanism and molecular basis for selenium/sulfur discrimination of selenocysteine lyase.
J.Biol.Chem., 285, 2010
|
|
5ZS9
 
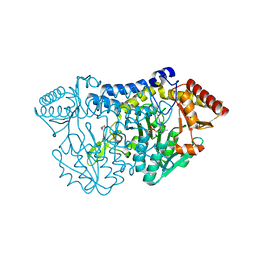 | |
5ZSS
 
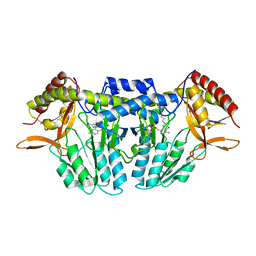 | |
5ZSR
 
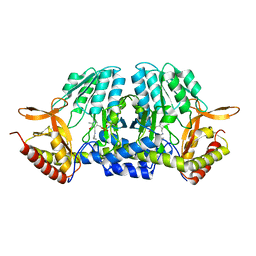 | |
5ZST
 
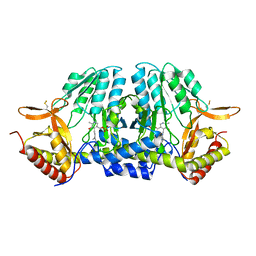 | |
5ZSQ
 
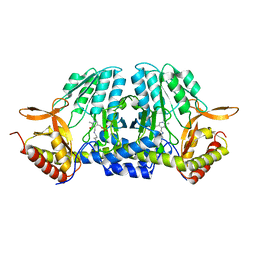 | |
2Z9V
 
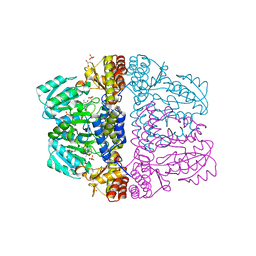 | | Crystal structure of pyridoxamine-pyruvate aminotransferase complexed with pyridoxamine | | Descriptor: | 4-(AMINOMETHYL)-5-(HYDROXYMETHYL)-2-METHYLPYRIDIN-3-OL, Aspartate aminotransferase, GLYCEROL, ... | | Authors: | Yoshikane, Y, Yokochi, N, Yamasaki, M, Mizutani, K, Ohnishi, K, Mikami, B, Hayashi, H, Yagi, T. | | Deposit date: | 2007-09-26 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti MAFF303099
J.Biol.Chem., 283, 2008
|
|
3CAI
 
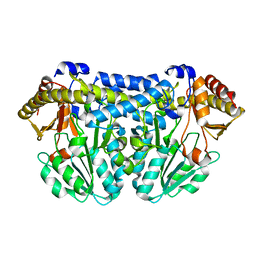 | | Crystal structure of Mycobacterium tuberculosis Rv3778c protein | | Descriptor: | POSSIBLE AMINOTRANSFERASE | | Authors: | Covarrubias, A.S, Larsson, A.M, Jones, T.A, Bergfors, T, Mowbray, S.L, Unge, T. | | Deposit date: | 2008-02-20 | | Release date: | 2008-03-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and biochemical function studies of Mycobacterium tuberculosis essential protein Rv3778c
To be published
|
|
2BI1
 
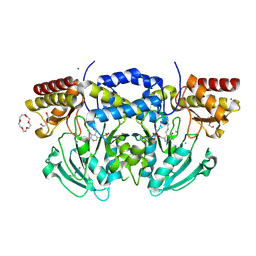 | | Radiation damage of the Schiff base in phosphoserine aminotransferase (structure B) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Dubnovitsky, A.P, Ravelli, R.B.G, Popov, A.N, Papageorgiou, A.C. | | Deposit date: | 2005-01-20 | | Release date: | 2005-05-19 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Strain Relief at the Active Site of Phosphoserine Aminotransferase Induced by Radiation Damage.
Protein Sci., 14, 2005
|
|
2BI5
 
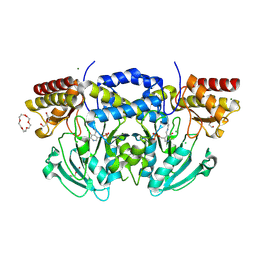 | | Radiation damage of the Schiff base in phosphoserine aminotransferase (structure E) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Dubnovitsky, A.P, Ravelli, R.B.G, Popov, A.N, Papageorgiou, A.C. | | Deposit date: | 2005-01-20 | | Release date: | 2005-05-19 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Strain Relief at the Active Site of Phosphoserine Aminotransferase Induced by Radiation Damage.
Protein Sci., 14, 2005
|
|
2BIA
 
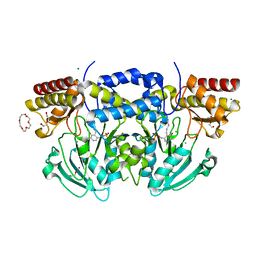 | | Radiation damage of the Schiff base in phosphoserine aminotransferase (structure G) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Dubnovitsky, A.P, Ravelli, R.B.G, Popov, A.N, Papageorgiou, A.C. | | Deposit date: | 2005-01-20 | | Release date: | 2005-05-19 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Strain Relief at the Active Site of Phosphoserine Aminotransferase Induced by Radiation Damage.
Protein Sci., 14, 2005
|
|
7CET
 
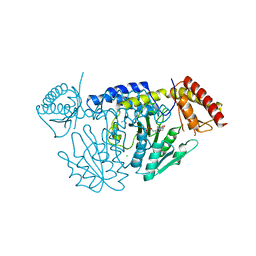 | | Crystal structure of D-cycloserine-bound form of cysteine desulfurase NifS from Helicobacter pylori | | Descriptor: | (5-hydroxy-6-methyl-4-{[(3-oxo-2,3-dihydro-1,2-oxazol-4-yl)amino]methyl}pyridin-3-yl)methyl dihydrogen phosphate, CHLORIDE ION, Cysteine desulfurase IscS, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2020-06-24 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 2022
|
|
7CEQ
 
 | |
7CER
 
 | | Crystal structure of D-cycloserine-bound form of cysteine desulfurase SufS H121A from Bacillus subtilis | | Descriptor: | Cysteine desulfurase SufS, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2020-06-24 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 2022
|
|
7CEP
 
 | | Crystal structure of L-cycloserine-bound form of cysteine desulfurase SufS from Bacillus subtilis | | Descriptor: | (5-hydroxy-6-methyl-4-{[(3-oxo-2,3-dihydro-1,2-oxazol-4-yl)amino]methyl}pyridin-3-yl)methyl dihydrogen phosphate, Cysteine desulfurase SufS, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nakamura, R, Yasuhiro, T, Fujishiro, T. | | Deposit date: | 2020-06-24 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 2022
|
|
7CEU
 
 | | Crystal structure of L-cycloserine-bound form of cysteine desulfurase NifS from Helicobacter pylori | | Descriptor: | (5-hydroxy-6-methyl-4-{[(3-oxo-2,3-dihydro-1,2-oxazol-4-yl)amino]methyl}pyridin-3-yl)methyl dihydrogen phosphate, Cysteine desulfurase IscS, ISOPROPYL ALCOHOL | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2020-06-24 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 2022
|
|
7CEO
 
 | |
7CES
 
 | | Crystal structure of L-cycloserine-bound form of cysteine desulfurase SufS H121A from Bacillus subtilis | | Descriptor: | (5-hydroxy-6-methyl-4-{[(3-oxo-2,3-dihydro-1,2-oxazol-4-yl)amino]methyl}pyridin-3-yl)methyl dihydrogen phosphate, Cysteine desulfurase SufS, DI(HYDROXYETHYL)ETHER | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2020-06-24 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 2022
|
|
2BI9
 
 | | Radiation damage of the Schiff base in phosphoserine aminotransferase (structure F) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Dubnovitsky, A.P, Ravelli, R.B.G, Popov, A.N, Papageorgiou, A.C. | | Deposit date: | 2005-01-20 | | Release date: | 2005-05-19 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Strain Relief at the Active Site of Phosphoserine Aminotransferase Induced by Radiation Damage.
Protein Sci., 14, 2005
|
|
2C0R
 
 | |
7E6A
 
 | | Crystal structure of cysteine desulfurase SufS C361A from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, Cysteine desulfurase SufS, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 289, 2022
|
|
2CH2
 
 | | Structure of the Anopheles gambiae 3-hydroxykynurenine transaminase in complex with inhibitor | | Descriptor: | 3-HYDROXYKYNURENINE TRANSAMINASE, 4-(2-AMINOPHENYL)-4-OXOBUTANOIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Rossi, F, Garavaglia, S, Giovenzana, G.B, Arca, B, Li, J, Rizzi, M. | | Deposit date: | 2006-03-10 | | Release date: | 2006-03-28 | | Last modified: | 2013-07-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Anopheles Gambiae 3-Hydroxykynurenine Transaminase
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
7E6E
 
 | | Crystal structure of PMP-bound form of cysteine desulfurase SufS R376A from Bacillus subtilis in D-cycloserine-inhibition | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Cysteine desulfurase SufS, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 289, 2022
|
|
7E6C
 
 | | Crystal structure of L-cycloserine-bound form of cysteine desulfurase SufS C361A from Bacillus subtilis | | Descriptor: | (5-hydroxy-6-methyl-4-{[(3-oxo-2,3-dihydro-1,2-oxazol-4-yl)amino]methyl}pyridin-3-yl)methyl dihydrogen phosphate, 1,2-ETHANEDIOL, Cysteine desulfurase SufS, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 289, 2022
|
|
7E6B
 
 | | Crystal structure of PMP-bound form of cysteine desulfurase SufS C361A from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Cysteine desulfurase SufS, ... | | Authors: | Nakamura, R, Takahashi, Y, Fujishiro, T. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Cycloserine enantiomers inhibit PLP-dependent cysteine desulfurase SufS via distinct mechanisms.
Febs J., 289, 2022
|
|
