7UOO
 
 | | Nucleoplasmic pre-60S intermediate of the Nog2 containing pre-rotation state | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 25S rRNA, 5.8S rRNA, ... | | Authors: | Sekulski, K, Cruz, V.E, Weirich, C.S, Erzberger, J.P. | | Deposit date: | 2022-04-13 | | Release date: | 2023-03-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | rRNA methylation by Spb1 regulates the GTPase activity of Nog2 during 60S ribosomal subunit assembly.
Nat Commun, 14, 2023
|
|
7V08
 
 | | Nucleoplasmic pre-60S intermediate of the Nog2 containing pre-rotation state from a Spb1 D52A suppressor 3 strain | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 25S rRNA, 5.8S rRNA, ... | | Authors: | Sekulski, K, Cruz, V.E, Weirich, C.S, Erzberger, J.P. | | Deposit date: | 2022-05-10 | | Release date: | 2023-03-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | rRNA methylation by Spb1 regulates the GTPase activity of Nog2 during 60S ribosomal subunit assembly.
Nat Commun, 14, 2023
|
|
7UQB
 
 | | Nucleoplasmic pre-60S intermediate of the Nog2 containing pre-rotation state from a SPB1-D52A strain with AlF4 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 25S rRNA, 5.8S rRNA, ... | | Authors: | Sekulski, K, Cruz, V.E, Weirich, C.S, Erzberger, J.P. | | Deposit date: | 2022-04-19 | | Release date: | 2023-03-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | rRNA methylation by Spb1 regulates the GTPase activity of Nog2 during 60S ribosomal subunit assembly.
Nat Commun, 14, 2023
|
|
7UQZ
 
 | | Nucleoplasmic pre-60S intermediate of the Nog2 containing pre-rotation state from a SPB1 D52A strain | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 25S rRNA, 5.8S rRNA, ... | | Authors: | Sekulski, K, Cruz, V.E, Weirich, C.S, Erzberger, J.P. | | Deposit date: | 2022-04-20 | | Release date: | 2023-03-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | rRNA methylation by Spb1 regulates the GTPase activity of Nog2 during 60S ribosomal subunit assembly.
Nat Commun, 14, 2023
|
|
8HFR
 
 | | NPC-trapped pre-60S particle | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Li, Z.Q, Chen, S.J, Sui, S.F. | | Deposit date: | 2022-11-12 | | Release date: | 2023-04-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Nuclear export of pre-60S particles through the nuclear pore complex.
Nature, 618, 2023
|
|
8V87
 
 | | 60S ribosome biogenesis intermediate (Dbp10 post-catalytic structure - Overall map) | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 25S ribosomal RNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Cruz, V.E, Weirich, C.S, Peddada, N, Erzberger, J.P. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | The DEAD-box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60S ribosome biogenesis.
Nat Commun, 15, 2024
|
|
7U0H
 
 | | State NE1 nucleolar 60S ribosome biogenesis intermediate - Overall model | | Descriptor: | 25S rRNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, 5.8S rRNA, ... | | Authors: | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | Deposit date: | 2022-02-18 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UG6
 
 | | Cryo-EM structure of pre-60S ribosomal subunit, unmethylated G2922 | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Yelland, J.N, Bravo, J.P.K, Black, J.J.B, Taylor, D.W, Johnson, A.W. | | Deposit date: | 2022-03-24 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A single 2'-O-methylation of ribosomal RNA gates assembly of a functional ribosome.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6YLG
 
 | |
7R7A
 
 | | State E1 nucleolar 60S ribosome biogenesis intermediate - Composite model | | Descriptor: | 25S rRNA, 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | Deposit date: | 2021-06-24 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7NAC
 
 | | State E2 nucleolar 60S ribosomal biogenesis intermediate - Composite model | | Descriptor: | 25S rRNA, 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | Deposit date: | 2021-06-21 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
3JCT
 
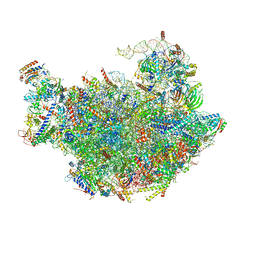 | | Cryo-em structure of eukaryotic pre-60S ribosomal subunits | | Descriptor: | 60S ribosomal protein L11-A, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | Authors: | Wu, S, Kumcuoglu, B, Yan, K.G, Brown, H, Zhang, Y.X, Tan, D, Gamalinda, M, Yuan, Y, Li, Z.F, Jakovljevic, J, Ma, C.Y, Lei, J.L, Dong, M.Q, Woolford Jr, J.L, Gao, N. | | Deposit date: | 2016-03-09 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes
Nature, 534, 2016
|
|
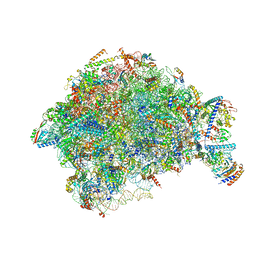
7OHQ
 
 | |
6YLH
 
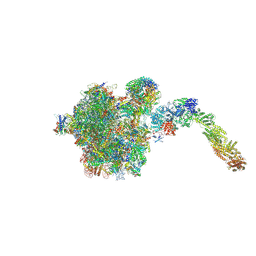 | |
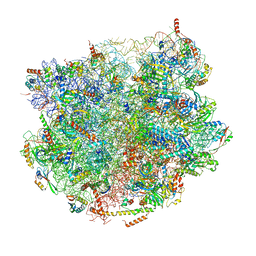
7BT6
 
 | |
7NAF
 
 | | State E2 nucleolar 60S ribosomal biogenesis intermediate - Spb1-MTD local model | | Descriptor: | 25S rRNA, 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | Deposit date: | 2021-06-21 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6M62
 
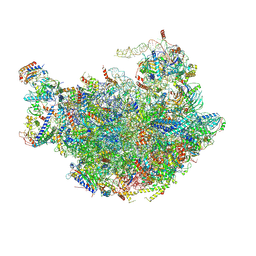 | |
7BTB
 
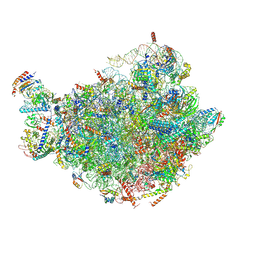 | |
6ELZ
 
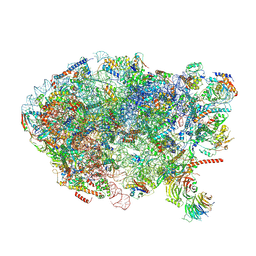 | | State E (TAP-Flag-Ytm1 E80A) - Visualizing the assembly pathway of nucleolar pre-60S ribosomes | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 25S ribosomal RNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Kater, L, Cheng, J, Barrio-Garcia, C, Hurt, E, Beckmann, R. | | Deposit date: | 2017-09-30 | | Release date: | 2017-12-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes.
Cell, 171, 2017
|
|
7OH3
 
 | |
6N8J
 
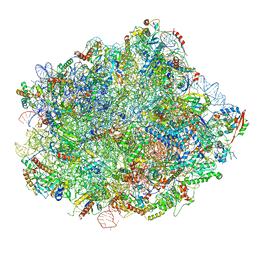 | | Cryo-EM structure of late nuclear (LN) pre-60S ribosomal subunit | | Descriptor: | 5.8S rRNA, 5S rRNA, 60S ribosomal protein L11-A, ... | | Authors: | Zhou, Y, Musalgaonkar, S, Johnson, A.W, Taylor, D.W. | | Deposit date: | 2018-11-29 | | Release date: | 2019-03-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Tightly-orchestrated rearrangements govern catalytic center assembly of the ribosome.
Nat Commun, 10, 2019
|
|
6EM1
 
 | | State C (Nsa1-TAP Flag-Ytm1) - Visualizing the assembly pathway of nucleolar pre-60S ribosomes | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 60S ribosomal protein L13-A, ... | | Authors: | Kater, L, Cheng, J, Barrio-Garcia, C, Hurt, E, Beckmann, R. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes.
Cell, 171, 2017
|
|
6N8K
 
 | | Cryo-EM structure of early cytoplasmic-immediate (ECI) pre-60S ribosomal subunit | | Descriptor: | 5.8S rRNA, 5S rRNA, 60S ribosomal export protein NMD3, ... | | Authors: | Zhou, Y, Musalgaonkar, S, Johnson, A.W, Taylor, D.W. | | Deposit date: | 2018-11-29 | | Release date: | 2019-03-13 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Tightly-orchestrated rearrangements govern catalytic center assembly of the ribosome.
Nat Commun, 10, 2019
|
|
6N8L
 
 | | Cryo-EM structure of early cytoplasmic-late (ECL) pre-60S ribosomal subunit | | Descriptor: | 5.8S rRNA, 5S rRNA, 60S ribosomal export protein NMD3, ... | | Authors: | Zhou, Y, Musalgaonkar, S, Johnson, A.W, Taylor, D.W. | | Deposit date: | 2018-11-29 | | Release date: | 2019-03-13 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Tightly-orchestrated rearrangements govern catalytic center assembly of the ribosome.
Nat Commun, 10, 2019
|
|
6YLY
 
 | | pre-60S State NE2 (TAP-Flag-Nop53) | | Descriptor: | 25S rRNA, 5.8S rRNA, 60S ribosomal protein L13-A, ... | | Authors: | Kater, L, Beckmann, R. | | Deposit date: | 2020-04-07 | | Release date: | 2020-07-29 | | Last modified: | 2020-09-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Construction of the Central Protuberance and L1 Stalk during 60S Subunit Biogenesis.
Mol.Cell, 79, 2020
|
|
