1GRN
 
 | | CRYSTAL STRUCTURE OF THE CDC42/CDC42GAP/ALF3 COMPLEX. | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Nassar, N, Hoffman, G.R, Clardy, J.C, Cerione, R.A. | | Deposit date: | 1998-07-30 | | Release date: | 1999-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Cdc42 bound to the active and catalytically compromised forms of Cdc42GAP.
Nat.Struct.Biol., 5, 1998
|
|
2NGR
 
 | | TRANSITION STATE COMPLEX FOR GTP HYDROLYSIS BY CDC42: COMPARISONS OF THE HIGH RESOLUTION STRUCTURES FOR CDC42 BOUND TO THE ACTIVE AND CATALYTICALLY COMPROMISED FORMS OF THE CDC42-GAP. | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Nassar, N, Hoffman, G, Clardy, J, Cerione, R. | | Deposit date: | 1998-07-31 | | Release date: | 1999-01-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Cdc42 bound to the active and catalytically compromised forms of Cdc42GAP.
Nat.Struct.Biol., 5, 1998
|
|
6W2E
 
 | | Structures of Capsid and Capsid-Associated Tegument Complex inside the Epstein-Barr Virus | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Liu, W, Cui, Y.X, Wang, C.Y, Li, Z.H, Gong, D.Y, Dai, X.H, Bi, G.Q, Sun, R, Zhou, Z.H. | | Deposit date: | 2020-03-05 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structures of capsid and capsid-associated tegument complex inside the Epstein-Barr virus.
Nat Microbiol, 5, 2020
|
|
7NWR
 
 | | Structure of BT1526, a myo-inositol-1-phosphate synthase | | Descriptor: | Inositol-3-phosphate synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | Basle, A, Tang, G, Marles-Wright, J, Campopiano, D. | | Deposit date: | 2021-03-17 | | Release date: | 2022-03-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of inositol lipid metabolism in gut-associated Bacteroidetes.
Nat Microbiol, 7, 2022
|
|
6DVK
 
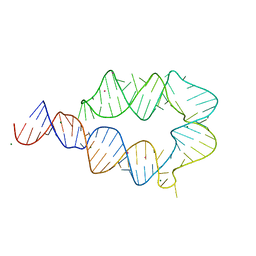 | | Computationally designed mini tetraloop-tetraloop receptor by the RNAMake program - construct 6 (miniTTR 6) | | Descriptor: | COBALT (II) ION, MAGNESIUM ION, RNA (95-MER) | | Authors: | Eiler, D.R, Yesselman, J.D, Costantino, D.A, Das, R, Kieft, J.S. | | Deposit date: | 2018-06-24 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Computational design of three-dimensional RNA structure and function.
Nat Nanotechnol, 14, 2019
|
|
8C8E
 
 | |
8CD1
 
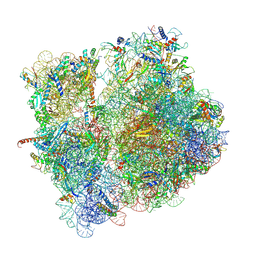 | | 70S-PHIKZ014 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Gerovac, M, Vogel, J. | | Deposit date: | 2023-01-29 | | Release date: | 2024-01-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Phage proteins target and co-opt host ribosomes immediately upon infection.
Nat Microbiol, 9, 2024
|
|
8HM2
 
 | |
8HM1
 
 | |
8HM3
 
 | | Complex of PPIase-BfUbb | | Descriptor: | GLYCEROL, MAGNESIUM ION, Peptidylprolyl isomerase, ... | | Authors: | Xu, J.H, Chen, Z, Gao, X. | | Deposit date: | 2022-12-02 | | Release date: | 2023-11-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Bacteroides fragilis ubiquitin homologue drives intraspecies bacterial competition in the gut microbiome.
Nat Microbiol, 9, 2024
|
|
6Q15
 
 | | Structure of the Salmonella SPI-1 injectisome needle complex | | Descriptor: | Lipoprotein PrgK, Protein InvG, Protein PrgH, ... | | Authors: | Hu, J, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2019-08-02 | | Release date: | 2019-10-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (5.15 Å) | | Cite: | T3S injectisome needle complex structures in four distinct states reveal the basis of membrane coupling and assembly.
Nat Microbiol, 4, 2019
|
|
8HM4
 
 | | Crystal structure of PPIase | | Descriptor: | Peptidylprolyl isomerase | | Authors: | Xu, J.H, Chen, Z, Gao, X. | | Deposit date: | 2022-12-02 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | Bacteroides fragilis ubiquitin homologue drives intraspecies bacterial competition in the gut microbiome.
Nat Microbiol, 9, 2024
|
|
6Q14
 
 | | Structure of the Salmonella SPI-1 injectisome NC-base | | Descriptor: | Lipoprotein PrgK, Protein InvG, Protein PrgH, ... | | Authors: | Hu, J, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2019-08-02 | | Release date: | 2019-10-23 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | T3S injectisome needle complex structures in four distinct states reveal the basis of membrane coupling and assembly.
Nat Microbiol, 4, 2019
|
|
6JIV
 
 | | SspE crystal structure | | Descriptor: | SspE protein | | Authors: | Bing, Y.Z, Yang, H.G. | | Deposit date: | 2019-02-23 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | SspABCD-SspE is a phosphorothioation-sensing bacterial defence system with broad anti-phage activities.
Nat Microbiol, 5, 2020
|
|
6Q64
 
 | | BT1044SeMet E190Q | | Descriptor: | Endoglycosidase | | Authors: | Basle, A, Paterson, N, Crouch, L, Bolam, D. | | Deposit date: | 2018-12-10 | | Release date: | 2019-05-08 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Complex N-glycan breakdown by gut Bacteroides involves an extensive enzymatic apparatus encoded by multiple co-regulated genetic loci.
Nat Microbiol, 4, 2019
|
|
8IGN
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with RAY1216 | | Descriptor: | (3~{S},3~{a}~{S},6~{a}~{R})-2-[(2~{S})-2-cyclohexyl-2-[2,2,2-tris(fluoranyl)ethanoylamino]ethanoyl]-~{N}-[(2~{S})-4-(cyclopentylamino)-3,4-bis(oxidanylidene)-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide, 3C-like proteinase nsp5 | | Authors: | Huang, X, Zhou, B, Xu, J, Yang, Z, Zhong, N, Xiong, X. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Preclinical evaluation of the SARS-CoV-2 M pro inhibitor RAY1216 shows improved pharmacokinetics compared with nirmatrelvir.
Nat Microbiol, 9, 2024
|
|
8IGO
 
 | | Crystal structure of apo SARS-CoV-2 main protease | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Huang, X, Zhou, B, Xu, J, Yang, Z, Zhong, N, Xiong, X. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Preclinical evaluation of the SARS-CoV-2 M pro inhibitor RAY1216 shows improved pharmacokinetics compared with nirmatrelvir.
Nat Microbiol, 9, 2024
|
|
6JUF
 
 | | SspB crystal structure | | Descriptor: | SspB protein | | Authors: | Liqiong, L, Yubing, Z. | | Deposit date: | 2019-04-13 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.587 Å) | | Cite: | SspABCD-SspE is a phosphorothioation-sensing bacterial defence system with broad anti-phage activities.
Nat Microbiol, 5, 2020
|
|
6Q2A
 
 | |
6JZJ
 
 | | Structure of FimA type-2 (FimA2) prepilin of the type V major fimbrium | | Descriptor: | Major fimbrium subunit FimA type-2, SULFATE ION | | Authors: | Okada, K, Shoji, M, Nakayam, K, Imada, K. | | Deposit date: | 2019-05-02 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of polymerized type V pilin reveals assembly mechanism involving protease-mediated strand exchange.
Nat Microbiol, 5, 2020
|
|
6JZK
 
 | | Structure of FimA type-1 (FimA1) prepilin of the type V major fimbrium | | Descriptor: | GLYCEROL, Major fimbrium subunit FimA type-1, S,R MESO-TARTARIC ACID, ... | | Authors: | Okada, K, Shoji, M, Nakayam, K, Imada, K. | | Deposit date: | 2019-05-02 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of polymerized type V pilin reveals assembly mechanism involving protease-mediated strand exchange.
Nat Microbiol, 5, 2020
|
|
6Q16
 
 | | Focussed refinement of InvGN0N1:PrgHK:SpaPQR:PrgIJ from Salmonella SPI-1 injectisome NC-base | | Descriptor: | Lipoprotein PrgK, Protein InvG, Protein PrgH, ... | | Authors: | Hu, J, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2019-08-02 | | Release date: | 2019-10-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | T3S injectisome needle complex structures in four distinct states reveal the basis of membrane coupling and assembly.
Nat Microbiol, 4, 2019
|
|
6Q63
 
 | | BT0459 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-hexosaminidase, CALCIUM ION, ... | | Authors: | Basle, A, Crouch, L, Bolam, D. | | Deposit date: | 2018-12-10 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Complex N-glycan breakdown by gut Bacteroides involves an extensive enzymatic apparatus encoded by multiple co-regulated genetic loci.
Nat Microbiol, 4, 2019
|
|
7SZI
 
 | | Cryo-EM structure of OmpK36-TraN mating pair stabilization proteins from carbapenem-resistant Klebsiella pneumoniae | | Descriptor: | OmpK36, TraN | | Authors: | Beltran, L.C, Seddon, C, Beis, K, Frankel, G, Egelman, E.H. | | Deposit date: | 2021-11-27 | | Release date: | 2022-06-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mating pair stabilization mediates bacterial conjugation species specificity.
Nat Microbiol, 7, 2022
|
|
7T12
 
 | | Hexameric HIV-1 (O-group) CA | | Descriptor: | Capsid protein p24 | | Authors: | Jacques, D.A, James, L.C. | | Deposit date: | 2021-12-01 | | Release date: | 2022-10-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Evasion of cGAS and TRIM5 defines pandemic HIV.
Nat Microbiol, 7, 2022
|
|
