1P3F
 
 | | Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants | | Descriptor: | Histone H2A, Histone H2B, Histone H3, ... | | Authors: | Muthurajan, U.M, Bao, Y, Forsberg, L.J, Edayathumangalam, R.S, Dyer, P.N, White, C.L, Luger, K. | | Deposit date: | 2003-04-17 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of histone Sin mutant nucleosomes reveal altered protein-DNA interactions
EMBO J., 23, 2004
|
|
1P3G
 
 | | Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants | | Descriptor: | Histone H2A, Histone H2B, Histone H3, ... | | Authors: | Muthurajan, U.M, Bao, Y, Forsberg, L.J, Edayathumangalam, R.S, Dyer, P.N, White, C.L, Luger, K. | | Deposit date: | 2003-04-17 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of histone Sin mutant nucleosomes reveal altered protein-DNA interactions
EMBO J., 23, 2004
|
|
1P3H
 
 | | Crystal Structure of the Mycobacterium tuberculosis chaperonin 10 tetradecamer | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 10 kDa chaperonin, CALCIUM ION | | Authors: | Roberts, M.M, Coker, A.R, Fossati, G, Mascagni, P, Coates, A.R.M, Wood, S.P, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-04-17 | | Release date: | 2003-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mycobacterium tuberculosis chaperonin 10 heptamers self-associate through their biologically active loops
J.BACTERIOL., 185, 2003
|
|
1P3I
 
 | | Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants | | Descriptor: | Histone H2A, Histone H2B, Histone H3, ... | | Authors: | Muthurajan, U.M, Bao, Y, Forsberg, L.J, Edayathumangalam, R.S, Dyer, P.N, White, C.L, Luger, K. | | Deposit date: | 2003-04-17 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of histone Sin mutant nucleosomes reveal altered protein-DNA interactions
EMBO J., 23, 2004
|
|
1P3J
 
 | | Adenylate Kinase from Bacillus subtilis | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bae, E, Phillips Jr, G.N. | | Deposit date: | 2003-04-17 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures and analysis of highly homologous psychrophilic, mesophilic, and thermophilic adenylate kinases.
J.Biol.Chem., 279, 2004
|
|
1P3K
 
 | | Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants | | Descriptor: | Histone H2A, Histone H2B, Histone H3, ... | | Authors: | Muthurajan, U.M, Bao, Y, Forsberg, L.J, Edayathumangalam, R.S, Dyer, P.N, White, C.L, Luger, K. | | Deposit date: | 2003-04-17 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of histone Sin mutant nucleosomes reveal altered protein-DNA interactions
EMBO J., 23, 2004
|
|
1P3L
 
 | | Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants | | Descriptor: | Histone H2A, Histone H2B, Histone H3, ... | | Authors: | Muthurajan, U.M, Bao, Y, Forsberg, L.J, Edayathumangalam, R.S, Dyer, P.N, White, C.L, Luger, K. | | Deposit date: | 2003-04-17 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of histone Sin mutant nucleosomes reveal altered protein-DNA interactions
EMBO J., 23, 2004
|
|
1P3M
 
 | | Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants | | Descriptor: | Histone H2A, Histone H2B, Histone H3, ... | | Authors: | Muthurajan, U.M, Bao, Y, Forsberg, L.J, Edayathumangalam, R.S, Dyer, P.N, White, C.L, Luger, K. | | Deposit date: | 2003-04-17 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of histone Sin mutant nucleosomes reveal altered protein-DNA interactions
EMBO J., 23, 2004
|
|
1P3N
 
 | | CORE REDESIGN BACK-REVERTANT I103V/CORE10 | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME, ... | | Authors: | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | Deposit date: | 2003-04-17 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Repacking the Core of T4 lysozyme by automated design
J.Mol.Biol., 332, 2003
|
|
1P3O
 
 | | Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants | | Descriptor: | Histone H2A, Histone H2B, Histone H3, ... | | Authors: | Muthurajan, U.M, Bao, Y, Forsberg, L.J, Edayathumangalam, R.S, Dyer, P.N, White, C.L, Luger, K. | | Deposit date: | 2003-04-17 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structures of histone Sin mutant nucleosomes reveal altered protein-DNA interactions
EMBO J., 23, 2004
|
|
1P3P
 
 | | Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants | | Descriptor: | Histone H2A, Histone H2B, Histone H3, ... | | Authors: | Muthurajan, U.M, Bao, Y, Forsberg, L.J, Edayathumangalam, R.S, Dyer, P.N, White, C.L, Luger, K. | | Deposit date: | 2003-04-17 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of histone Sin mutant nucleosomes reveal altered protein-DNA interactions
EMBO J., 23, 2004
|
|
1P3Q
 
 | | Mechanism of Ubiquitin Recognition by the CUE Domain of VPS9 | | Descriptor: | Ubiquitin, Vacuolar protein sorting-associated protein VPS9 | | Authors: | Prag, G, Misra, S, Jones, E.A, Ghirlando, R, Davies, B.A, Horazdovsky, B.F, Hurley, J.H. | | Deposit date: | 2003-04-18 | | Release date: | 2003-06-24 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of Ubiquitin Recognition by the CUE Domain of Vps9p.
Cell(Cambridge,Mass.), 113, 2003
|
|
1P3R
 
 | | Crystal structure of the phosphotyrosin binding domain(PTB) of mouse Disabled 1(Dab1) | | Descriptor: | Disabled homolog 2 | | Authors: | Yun, M, Keshvara, L, Park, C.G, Zhang, Y.M, Dickerson, J.B, Zheng, J, Rock, C.O, Curran, T, Park, H.W. | | Deposit date: | 2003-04-18 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the Dab homology domains of mouse disabled 1 and 2.
J.Biol.Chem., 278, 2003
|
|
1P3T
 
 | | Crystal Structures of the NO-and CO-Bound Heme Oxygenase From Neisseria Meningitidis: Implications for Oxygen Activation | | Descriptor: | Heme oxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Friedman, J, Lad, L, Deshmukh, R, Li, H, Wilks, A, Poulos, T.L. | | Deposit date: | 2003-04-18 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the NO- and CO-bound heme oxygenase from Neisseriae meningitidis. Implications for O2 activation
J.Biol.Chem., 278, 2003
|
|
1P3U
 
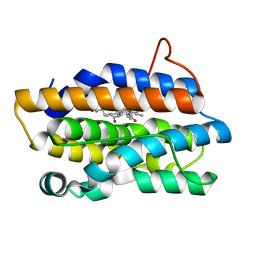 | | Crystal Structures of the NO-and CO-Bound Heme Oxygenase From Neisseria Meningitidis: Implications for Oxygen Activation | | Descriptor: | Heme oxygenase 1, NITRIC OXIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Friedman, J, Lad, L, Deshmukh, R, Li, H, Wilks, A, Poulos, T.L. | | Deposit date: | 2003-04-18 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of the NO- and CO-bound heme oxygenase from Neisseriae meningitidis. Implications for O2 activation
J.Biol.Chem., 278, 2003
|
|
1P3V
 
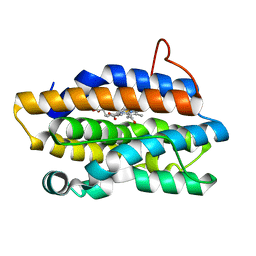 | | Crystal Structures of the NO-and CO-Bound Heme Oxygenase From Neisseria Meningitidis: Implications for Oxygen Activation | | Descriptor: | CARBON MONOXIDE, Heme oxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Friedman, J, Lad, L, Deshmukh, R, Li, H, Wilks, A, Poulos, T.L. | | Deposit date: | 2003-04-18 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of the NO- and CO-bound heme oxygenase from Neisseriae meningitidis. Implications for O2 activation
J.Biol.Chem., 278, 2003
|
|
1P3W
 
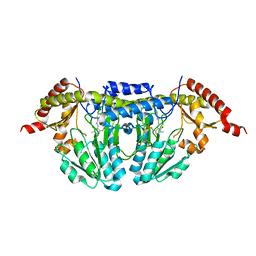 | |
1P3X
 
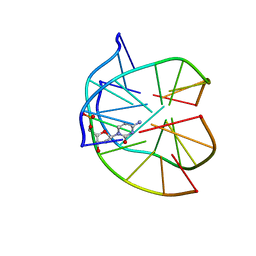 | | INTRAMOLECULAR DNA TRIPLEX WITH 1-PROPYNYL DEOXYURIDINE IN THE THIRD STRAND, NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*(PDU)P*CP*(PDU)P*(DCM)P*(PDU)P*CP*(PDU)P*(PDU))-3'), DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*A)-3'), DNA (5'-D(*TP*TP*CP*TP*CP*TP*CP*T)-3') | | Authors: | Phipps, A.K, Tarkoy, M, Schultze, P, Feigon, J. | | Deposit date: | 1998-02-05 | | Release date: | 1998-05-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an intramolecular DNA triplex containing 5-(1-propynyl)-2'-deoxyuridine residues in the third strand.
Biochemistry, 37, 1998
|
|
1P3Y
 
 | | MrsD from Bacillus sp. HIL-Y85/54728 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MrsD protein | | Authors: | Blaesse, M, Kupke, T, Huber, R, Steinbacher, S. | | Deposit date: | 2003-04-19 | | Release date: | 2003-08-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structure of MrsD, an FAD-binding protein of the HFCD family.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1P42
 
 | | Crystal structure of Aquifex aeolicus LpxC Deacetylase (Zinc-Inhibited Form) | | Descriptor: | MYRISTIC ACID, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Whittington, D.A, Rusche, K.M, Shin, H, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2003-04-21 | | Release date: | 2003-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of LpxC, a Zinc-Dependent Deacetylase Essential for Endotoxin Biosynthesis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1P43
 
 | | REVERSE PROTONATION IS THE KEY TO GENERAL ACID-BASE CATALYSIS IN ENOLASE | | Descriptor: | 2-PHOSPHOGLYCERIC ACID, Enolase 1, MAGNESIUM ION | | Authors: | Sims, P.A, Larsen, T.M, Poyner, R.R, Cleland, W.W, Reed, G.H. | | Deposit date: | 2003-04-21 | | Release date: | 2003-11-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reverse protonation is the key to general acid-base catalysis in enolase
Biochemistry, 42, 2003
|
|
1P44
 
 | | Targeting tuberculosis and malaria through inhibition of enoyl reductase: compound activity and structural data | | Descriptor: | 5-{[4-(9H-FLUOREN-9-YL)PIPERAZIN-1-YL]CARBONYL}-1H-INDOLE, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kuo, M.R, Morbidoni, H.R, Alland, D, Sneddon, S.F, Gourlie, B.B, Staveski, M.M, Leonard, M, Gregory, J.S, Janjigian, A.D, Yee, C, Musser, J.M, Kreiswirth, B, Iwamoto, H, Perozzo, R, Jacobs Jr, W.R, Sacchettini, J.C, Fidock, D.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-04-21 | | Release date: | 2003-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Targeting tuberculosis and malaria through inhibition of Enoyl reductase: compound activity and structural data.
J.Biol.Chem., 278, 2003
|
|
1P45
 
 | | Targeting tuberculosis and malaria through inhibition of enoyl reductase: compound activity and structural data | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Kuo, M.R, Morbidoni, H.R, Alland, D, Sneddon, S.F, Gourlie, B.B, Staveski, M.M, Leonard, M, Gregory, J.S, Janjigian, A.D, Yee, C, Musser, J.M, Kreiswirth, B.N, Iwamoto, H, Perozzo, R, Jacobs Jr, W.R, Sacchettini, J.C, Fidock, D.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-04-21 | | Release date: | 2003-09-16 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Targeting tuberculosis and malaria through inhibition of Enoyl reductase: compound activity and structural data.
J.Biol.Chem., 278, 2003
|
|
1P46
 
 | | T4 lysozyme core repacking mutant M106I/TA | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME, ... | | Authors: | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | Deposit date: | 2003-04-21 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Repacking the Core of T4 lysozyme by automated design
J.Mol.Biol., 332, 2003
|
|
1P47
 
 | | Crystal Structure of tandem Zif268 molecules complexed to DNA | | Descriptor: | 5'-D(*CP*AP*CP*GP*CP*CP*CP*AP*CP*GP*CP*CP*GP*CP*CP*CP*AP*CP*GP*CP*CP*A)-3', 5'-D(*GP*TP*GP*GP*CP*GP*TP*GP*GP*GP*CP*GP*GP*CP*GP*TP*GP*GP*GP*CP*GP*T)-3', Early growth response protein 1, ... | | Authors: | Peisach, E, Pabo, C.O. | | Deposit date: | 2003-04-21 | | Release date: | 2003-06-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Constraints for Zinc Finger Linker Design as Inferred from X-ray Crystal Structure of Tandem Zif268-DNA Complexes
J.Mol.Biol., 330, 2003
|
|
