1NTP
 
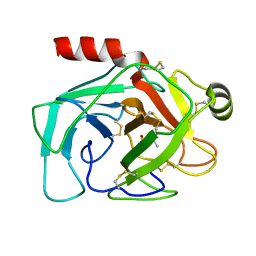 | |
1NTQ
 
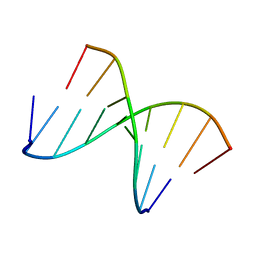 | | 5'(dCCUCCUU)3':3'(rAGGAGGAAA)5' | | Descriptor: | 5'-D(*CP*CP*UP*CP*CP*UP*U)-3', 5'-R(*AP*AP*AP*GP*GP*AP*GP*GP*A)-3' | | Authors: | Znosko, B.M, Barnes III, T.W, Krugh, T.R, Turner, D.H. | | Deposit date: | 2003-01-30 | | Release date: | 2003-06-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Studies of DNA Single Strands and DNA:RNA Hybrids With and Without 1-Propynylation at C5 of Oligopyrimidines
J.Am.Chem.Soc., 125, 2003
|
|
1NTR
 
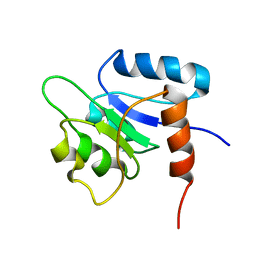 | | SOLUTION STRUCTURE OF THE N-TERMINAL RECEIVER DOMAIN OF NTRC | | Descriptor: | NTRC RECEIVER DOMAIN | | Authors: | Volkman, B.F, Nohaile, M.J, Amy, N.K, Kustu, S, Wemmer, D.E. | | Deposit date: | 1994-09-16 | | Release date: | 1995-01-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the N-terminal receiver domain of NTRC.
Biochemistry, 34, 1995
|
|
1NTS
 
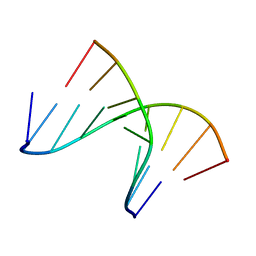 | | 5'(dCCPUPCPCPUPUP)3':3'(rAGGAGGAAA)5', where P=propynyl | | Descriptor: | 5'-D(*(5PC)P*(5PC)P*(PDU)P*(5PC)P*(5PC)P*(PDU)P*(PDU))-3', 5'-R(*AP*AP*AP*GP*GP*AP*GP*GP*A)-3' | | Authors: | Znosko, B.M, Barnes III, T.W, Krugh, T.R, Turner, D.H. | | Deposit date: | 2003-01-30 | | Release date: | 2003-06-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Studies of DNA Single Strands and DNA:RNA Hybrids With and Without 1-Propynylation at C5 of Oligopyrimidines
J.Am.Chem.Soc., 125, 2003
|
|
1NTT
 
 | | 5'(dCPCPUPCPCPUPUP)3':(rAGGAGGAAA)5', where P=propynyl | | Descriptor: | 5'-D(*CP*(5PC)P*(PDU)P*(5PC)P*(5PC)P*(PDU)P*(PDU))-3', 5'-R(*AP*AP*AP*GP*GP*AP*GP*GP*A)-3' | | Authors: | Znosko, B.M, Barnes III, T.W, Krugh, T.R, Turner, D.H. | | Deposit date: | 2003-01-30 | | Release date: | 2003-06-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Studies of DNA Single Strands and DNA:RNA Hybrids With and Without 1-Propynylation at C5 of Oligopyrimidines
J.Am.Chem.Soc., 125, 2003
|
|
1NTV
 
 | | Crystal Structure of the Disabled-1 (Dab1) PTB domain-ApoER2 peptide complex | | Descriptor: | Apolipoprotein E Receptor-2 peptide, Disabled homolog 1, PHOSPHATE ION | | Authors: | Stolt, P.C, Jeon, H, Song, H.K, Herz, J, Eck, M.J, Blacklow, S.C. | | Deposit date: | 2003-01-30 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Origins of Peptide Selectivity and Phosphoinositide Binding Revealed by Structures of Disabled-1 PTB Domain Complexes
Structure, 11, 2003
|
|
1NTX
 
 | |
1NTY
 
 | |
1NTZ
 
 | | Crystal Structure of Mitochondrial Cytochrome bc1 Complex Bound with Ubiquinone | | Descriptor: | Cytochrome b, FE2/S2 (INORGANIC) CLUSTER, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Gao, X, Wen, X, Esser, L, Quinn, B, Yu, L, Yu, C.-A, Xia, D. | | Deposit date: | 2003-01-30 | | Release date: | 2003-10-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the quinone reduction in the bc(1) complex: a comparative analysis of crystal structures of mitochondrial cytochrome bc(1) with bound substrate and inhibitors at the Q(i) site
Biochemistry, 42, 2003
|
|
1NU0
 
 | | Structure of the double mutant (L6M; F134M, SeMet form) of yqgF from Escherichia coli, a hypothetical protein | | Descriptor: | Hypothetical protein yqgF, SULFATE ION | | Authors: | Galkin, A, Sarikaya, E, Krajewski, W, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-01-30 | | Release date: | 2004-03-02 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of yqgF from Escherichia coli, a hypothetical protein
To be Published
|
|
1NU1
 
 | | Crystal Structure of Mitochondrial Cytochrome bc1 Complexed with 2-nonyl-4-hydroxyquinoline N-oxide (NQNO) | | Descriptor: | 2-NONYL-4-HYDROXYQUINOLINE N-OXIDE, Cytochrome b, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Gao, X, Wen, X, Esser, L, Quinn, B, Yu, L, Yu, C.-A, Xia, D. | | Deposit date: | 2003-01-30 | | Release date: | 2003-10-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the quinone reduction in the bc(1) complex: a comparative analysis of crystal structures of mitochondrial cytochrome bc(1) with bound substrate and inhibitors at the Q(i) site
Biochemistry, 42, 2003
|
|
1NU2
 
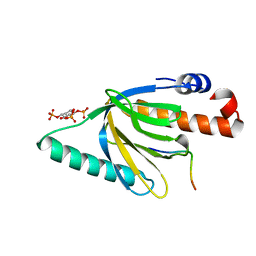 | | Crystal structure of the murine Disabled-1 (Dab1) PTB domain-ApoER2 peptide-PI-4,5P2 ternary complex | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Disabled homolog 1, peptide derived from murine Apolipoprotein E Receptor-2 | | Authors: | Stolt, P.C, Jeon, H, Song, H.K, Herz, J, Eck, M.J, Blacklow, S.C. | | Deposit date: | 2003-01-30 | | Release date: | 2003-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Origins of Peptide Selectivity and Phosphoinositide Binding Revealed by Structures of Disabled-1 PTB Domain Complexes
Structure, 11, 2003
|
|
1NU3
 
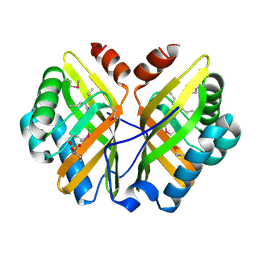 | | Limonene-1,2-epoxide hydrolase in complex with valpromide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-PROPYLPENTANAMIDE, limonene-1,2-epoxide hydrolase | | Authors: | Arand, M, Hallberg, B.M, Zou, J, Bergfors, T, Oesch, F, van der Werf, M.J, de Bont, J.A.M, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2003-01-30 | | Release date: | 2003-06-10 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of Rhodococcus erythropolis limonene-1,2-epoxide hydrolase reveals a novel active site
EMBO J., 22, 2003
|
|
1NU4
 
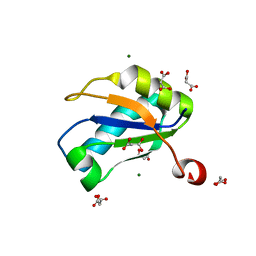 | | U1A RNA binding domain at 1.8 angstrom resolution reveals a pre-organized C-terminal helix | | Descriptor: | MAGNESIUM ION, MALONIC ACID, U1A RNA binding domain | | Authors: | Rupert, P.B, Xiao, H, Ferre-D'Amare, A.R. | | Deposit date: | 2003-01-30 | | Release date: | 2003-02-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | U1A RNA-binding domain at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1NU5
 
 | |
1NU6
 
 | | Crystal structure of human Dipeptidyl Peptidase IV (DPP-IV) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase IV, MERCURY (II) ION | | Authors: | Hennig, M, Stihle, M, Thoma, R, Ruf, A. | | Deposit date: | 2003-01-31 | | Release date: | 2003-08-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Proline-Specific Exopeptidase Activity as Observed in Human Dipeptidyl Peptidase-IV.
Structure, 11, 2003
|
|
1NU7
 
 | | Staphylocoagulase-Thrombin Complex | | Descriptor: | IMIDAZOLE, MERCURY (II) ION, N-(sulfanylacetyl)-D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Friedrich, R, Bode, W, Fuentes-Prior, P, Panizzi, P, Bock, P.E. | | Deposit date: | 2003-01-31 | | Release date: | 2003-10-07 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Staphylocoagulase is a prototype for the mechanism of cofactor-induced zymogen activation
NATURE, 425, 2003
|
|
1NU8
 
 | | Crystal structure of human dipeptidyl peptidase IV (DPP-IV) in complex with Diprotin A (IPI) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-mer peptide, Dipeptidyl peptidase IV | | Authors: | Thoma, R, Loeffler, B, Stihle, M, Huber, W, Ruf, A, Hennig, M. | | Deposit date: | 2003-01-31 | | Release date: | 2003-08-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of Proline-Specific Exopeptidase Activity as Observed in Human Dipeptidyl Peptidase-IV.
Structure, 11, 2003
|
|
1NU9
 
 | | Staphylocoagulase-Prethrombin-2 complex | | Descriptor: | IMIDAZOLE, MERCURY (II) ION, N-(sulfanylacetyl)-D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Friedrich, R, Bode, W, Fuentes-Prior, P, Panizzi, P, Bock, P.E. | | Deposit date: | 2003-01-31 | | Release date: | 2003-10-07 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Staphylocoagulase is a prototype for the mechanism of cofactor-induced zymogen activation
NATURE, 425, 2003
|
|
1NUA
 
 | | Crystal structure of 3-dehydroquinate synthase (DHQS) in complex with ZN2+ | | Descriptor: | 3-DEHYDROQUINATE SYNTHASE, CHLORIDE ION, ZINC ION | | Authors: | Nichols, C.E, Ren, J, Lamb, H.K, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2003-01-31 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Ligand-induced Conformational Changes and a Mechanism for Domain Closure in Aspergillus nidulans Dehydroquinate Synthase
J.MOL.BIOL., 327, 2003
|
|
1NUB
 
 | | HELIX C DELETION MUTANT OF BM-40 FS-EC DOMAIN PAIR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BASEMENT MEMBRANE PROTEIN BM-40, CALCIUM ION | | Authors: | Hohenester, E, Sasaki, T, Timpl, R. | | Deposit date: | 1997-12-05 | | Release date: | 1998-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure and mapping by site-directed mutagenesis of the collagen-binding epitope of an activated form of BM-40/SPARC/osteonectin.
EMBO J., 17, 1998
|
|
1NUC
 
 | | STAPHYLOCOCCAL NUCLEASE, V23C VARIANT | | Descriptor: | CALCIUM ION, STAPHYLOCOCCAL NUCLEASE, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Wynn, R, Harkins, P.C, Richards, F.M, Fox, R.O. | | Deposit date: | 1997-02-15 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mobile unnatural amino acid side chains in the core of staphylococcal nuclease.
Protein Sci., 5, 1996
|
|
1NUD
 
 | |
1NUE
 
 | | X-RAY STRUCTURE OF NM23 HUMAN NUCLEOSIDE DIPHOSPHATE KINASE B COMPLEXED WITH GDP AT 2 ANGSTROMS RESOLUTION | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Morera, S, Lacombe, M.-L, Yingwu, X, Lebras, G, Janin, J. | | Deposit date: | 1995-10-06 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of human nucleoside diphosphate kinase B complexed with GDP at 2 A resolution.
Structure, 3, 1995
|
|
1NUF
 
 | |
