1M0L
 
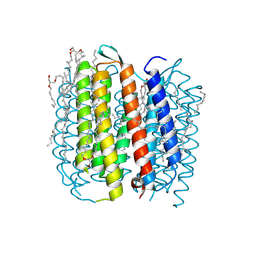 | | BACTERIORHODOPSIN/LIPID COMPLEX AT 1.47 A RESOLUTION | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, 2,10,23-TRIMETHYL-TETRACOSANE, BACTERIORHODOPSIN, ... | | Authors: | Lanyi, J.K. | | Deposit date: | 2002-06-13 | | Release date: | 2002-09-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystallographic structure of the K intermediate of bacteriorhodopsin: conservation of free energy after photoisomerization of the retinal.
J.Mol.Biol., 321, 2002
|
|
1M0M
 
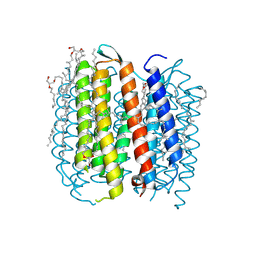 | | BACTERIORHODOPSIN M1 INTERMEDIATE AT 1.43 A RESOLUTION | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, 2,10,23-TRIMETHYL-TETRACOSANE, BACTERIORHODOPSIN, ... | | Authors: | Lanyi, J.K. | | Deposit date: | 2002-06-13 | | Release date: | 2002-09-11 | | Last modified: | 2015-09-09 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystallographic structure of the retinal and the protein after deprotonation of the Schiff base: the switch in the bacteriorhodopsin photocycle.
J.Mol.Biol., 321, 2002
|
|
1M0N
 
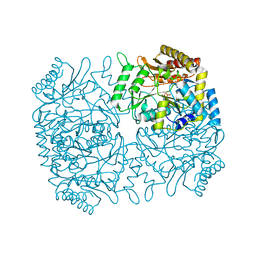 | | Structure of Dialkylglycine Decarboxylase Complexed with 1-Aminocyclopentanephosphonate | | Descriptor: | 1-[((1E)-{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYLENE)AMINO]CYCLOPENTYLPHOSPHONIC ACID, 2,2-Dialkylglycine decarboxylase, POTASSIUM ION, ... | | Authors: | Liu, W, Rogers, C.J, Fisher, A.J, Toney, M.D. | | Deposit date: | 2002-06-13 | | Release date: | 2002-10-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Aminophosphonate Inhibitors of Dialkylglycine Decarboxylase: Structural Basis for Slow Binding Inhibition
Biochemistry, 41, 2002
|
|
1M0O
 
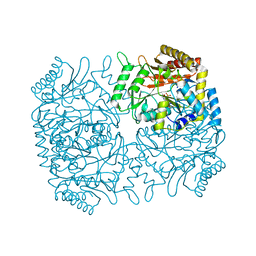 | | Structure of Dialkylglycine Decarboxylase Complexed with 1-Amino-1-methylpropanephosphonate | | Descriptor: | (1R)-1-[((1E)-{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYLENE)AMINO]-1-METHYLPROPYLPHOSPHONIC ACID, 2,2-Dialkylglycine decarboxylase, POTASSIUM ION, ... | | Authors: | Liu, W, Rogers, C.J, Fisher, A.J, Toney, M.D. | | Deposit date: | 2002-06-13 | | Release date: | 2002-10-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Aminophosphonate Inhibitors of Dialkylglycine Decarboxylase: Structural Basis for Slow Binding Inhibition
Biochemistry, 41, 2002
|
|
1M0P
 
 | | Structure of Dialkylglycine Decarboxylase Complexed with 1-Amino-1-phenylethanephosphonate | | Descriptor: | (1R)-1-[((1E)-{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYLENE)AMINO]-1-PHENYLETHYLPHOSPHONIC ACID, 2,2-Dialkylglycine Decarboxylase, POTASSIUM ION, ... | | Authors: | Liu, W, Rogers, C.J, Fisher, A.J, Toney, M.D. | | Deposit date: | 2002-06-13 | | Release date: | 2002-10-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Aminophosphonate Inhibitors of Dialkylglycine Decarboxylase: Structural Basis for Slow Binding Inhibition
Biochemistry, 41, 2002
|
|
1M0Q
 
 | | Structure of Dialkylglycine Decarboxylase Complexed with S-1-aminoethanephosphonate | | Descriptor: | (1S)-1-[((1E)-{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYLENE)AMINO]ETHYLPHOSPHONIC ACID, 2,2-Dialkylglycine Decarboxylase, POTASSIUM ION, ... | | Authors: | Liu, W, Rogers, C.J, Fisher, A.J, Toney, M.D. | | Deposit date: | 2002-06-13 | | Release date: | 2002-10-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aminophosphonate Inhibitors of Dialkylglycine Decarboxylase: Structural Basis for Slow Binding Inhibition
Biochemistry, 41, 2002
|
|
1M0S
 
 | | NORTHEAST STRUCTURAL GENOMICS CONSORTIUM (NESG ID IR21) | | Descriptor: | CITRIC ACID, Ribose-5-Phosphate Isomerase A | | Authors: | Das, K, Xiao, R, Acton, T, Montelione, G, Arnold, E, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-06-14 | | Release date: | 2002-09-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D-RIBOSE-5-PHOSPHATE ISOMERASE, IR21
TO BE PUBLISHED
|
|
1M0T
 
 | |
1M0U
 
 | | Crystal Structure of the Drosophila Glutathione S-transferase-2 in Complex with Glutathione | | Descriptor: | GLUTATHIONE, GST2 gene product, SULFATE ION | | Authors: | Agianian, B, Tucker, P.A, Schouten, A, Leonard, K, Bullard, B, Gros, P. | | Deposit date: | 2002-06-14 | | Release date: | 2003-02-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of a Drosophila Sigma Class Glutathione S-transferase Reveals a Novel
Active Site Topography Suited for Lipid Peroxidation Products
J.Mol.Biol., 326, 2003
|
|
1M0V
 
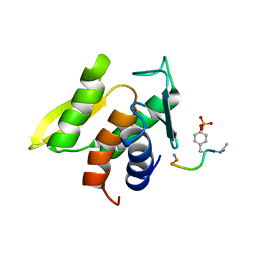 | | NMR STRUCTURE OF THE TYPE III SECRETORY DOMAIN OF YERSINIA YOPH COMPLEXED WITH THE SKAP-HOM PHOSPHO-PEPTIDE N-acetyl-DEpYDDPF-NH2 | | Descriptor: | PROTEIN-TYROSINE PHOSPHATASE YOPH, SKAP55 homologue | | Authors: | Khandelwal, P, Keliikuli, K, Smith, C.L, Saper, M.A, Zuiderweg, E.R.P. | | Deposit date: | 2002-06-14 | | Release date: | 2002-07-24 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and phosphopeptide binding to the N-terminal domain of Yersinia YopH: comparison with a crystal structure
Biochemistry, 41, 2002
|
|
1M0W
 
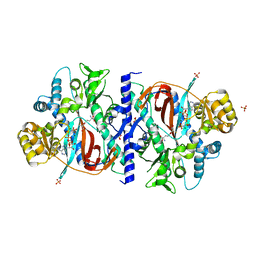 | | Yeast Glutathione Synthase Bound to gamma-glutamyl-cysteine, AMP-PNP and 2 Magnesium Ions | | Descriptor: | GAMMA-GLUTAMYLCYSTEINE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Gogos, A, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2002-06-14 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Large Conformational Changes in the Catalytic Cycle of Glutathione Synthase
Structure, 10, 2002
|
|
1M0Z
 
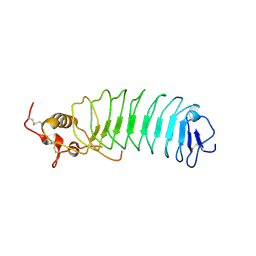 | | Crystal Structure of the von Willebrand Factor Binding Domain of Glycoprotein Ib alpha | | Descriptor: | Glycoprotein Ib alpha | | Authors: | Huizinga, E.G, Tsuji, S, Romijn, R.A.P, Schiphorst, M.E, de Groot, P.G, Sixma, J.J, Gros, P. | | Deposit date: | 2002-06-16 | | Release date: | 2002-08-28 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of glycoprotein Ibalpha and its complex with von Willebrand factor A1 domain.
Science, 297, 2002
|
|
1M10
 
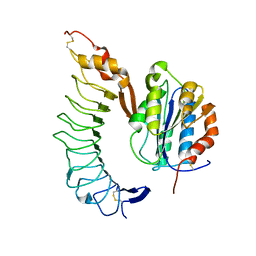 | | Crystal structure of the complex of Glycoprotein Ib alpha and the von Willebrand Factor A1 Domain | | Descriptor: | Glycoprotein Ib alpha, von Willebrand Factor | | Authors: | Huizinga, E.G, Tsuji, S, Romijn, R.A.P, Schiphorst, M.E, de Groot, P.G, Sixma, J.J, Gros, P. | | Deposit date: | 2002-06-16 | | Release date: | 2002-08-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of glycoprotein Ibalpha and its complex with von Willebrand factor A1 domain.
Science, 297, 2002
|
|
1M11
 
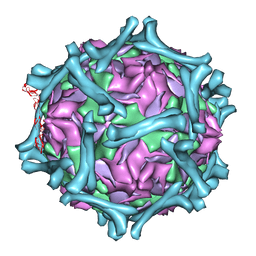 | | structural model of human decay-accelerating factor bound to echovirus 7 from cryo-electron microscopy | | Descriptor: | COAT PROTEIN VP1, COAT PROTEIN VP2, COAT PROTEIN VP3, ... | | Authors: | He, Y, Lin, F, Chipman, P.R, Bator, C.M, Baker, T.S, Shoham, M, Kuhn, R.J, Medof, M.E, Rossmann, M.G. | | Deposit date: | 2002-06-17 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Structure of decay-accelerating factor bound to echovirus 7: a virus-receptor complex.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1M12
 
 | |
1M13
 
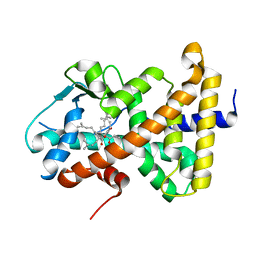 | | Crystal Structure of the Human Pregane X Receptor Ligand Binding Domain in Complex with Hyperforin, a Constituent of St. John's Wort | | Descriptor: | 4-HYDROXY-5-ISOBUTYRYL-6-METHYL-1,3,7-TRIS-(3-METHYL-BUT-2-ENYL)-6-(4-METHYL-PENT-3-ENYL)-BICYCLO[3.3.1]NON-3-ENE-2,9-DIONE, Orphan Nuclear Receptor PXR | | Authors: | Watkins, R.E, Maglich, J.M, Moore, L.B, Wisely, G.B, Noble, S.M, Davis-Searles, P.R, Lambert, M.H, Kliewer, S.A, Redinbo, M.R. | | Deposit date: | 2002-06-17 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 2.1 A Crystal Structure of Human PXR in Complex with the St. John's Wort Compound Hyperforin
Biochemistry, 42, 2003
|
|
1M14
 
 | |
1M15
 
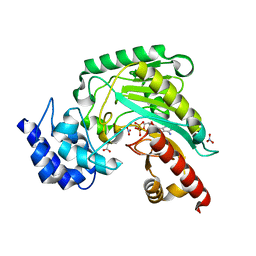 | | Transition state structure of arginine kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE, MAGNESIUM ION, ... | | Authors: | Yousef, M.S, Fabiola, F, Gattis, J.L, Somasundaram, T, Chapman, M.S. | | Deposit date: | 2002-06-17 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Refinement of the arginine kinase transition-state analogue complex at 1.2 A resolution: mechanistic insights.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1M16
 
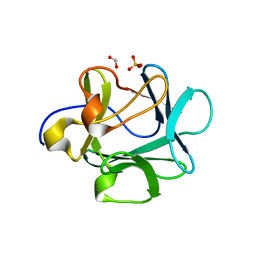 | | Human Acidic Fibroblast Growth Factor. 141 Amino Acid Form with Amino Terminal His Tag and Leu 44 Replaced with Phe (L44F), Leu 73 Replaced with Val (L73V), Val 109 Replaced with Leu (V109L) and Cys 117 Replaced with Val (C117V). | | Descriptor: | FORMIC ACID, SULFATE ION, acidic fibroblast growth factor | | Authors: | Brych, S.R, Kim, J, Spielmann, G.L, Logan, T.M, Blaber, M. | | Deposit date: | 2002-06-17 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Accommodation of a highly symmetric core within a symmetric protein superfold
Protein Sci., 12, 2003
|
|
1M17
 
 | |
1M18
 
 | | LIGAND BINDING ALTERS THE STRUCTURE AND DYNAMICS OF NUCLEOSOMAL DNA | | Descriptor: | Histone H2A.1, Histone H2B.1, Histone H3.2, ... | | Authors: | Suto, R.K, Edayathumangalam, R.S, White, C.L, Melander, C, Gottesfeld, J.M, Dervan, P.B, Luger, K. | | Deposit date: | 2002-06-18 | | Release date: | 2003-02-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structures of Nucleosome Core Particles in Complex with Minor Groove DNA-binding Ligands
J.Mol.Biol., 326, 2003
|
|
1M19
 
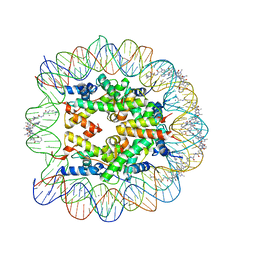 | | LIGAND BINDING ALTERS THE STRUCTURE AND DYNAMICS OF NUCLEOSOMAL DNA | | Descriptor: | 3-AMINO-(DIMETHYLPROPYLAMINE), 4-AMINO-(1-METHYLIMIDAZOLE)-2-CARBOXYLIC ACID, 4-AMINO-(1-METHYLPYRROLE)-2-CARBOXYLIC ACID, ... | | Authors: | Suto, R.K, Edayathumangalam, R.S, White, C.L, Melander, C, Gottesfeld, J.M, Dervan, P.B, Luger, K. | | Deposit date: | 2002-06-18 | | Release date: | 2003-02-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Nucleosome Core Particles in Complex with Minor Groove DNA-binding Ligands
J.Mol.Biol., 326, 2003
|
|
1M1A
 
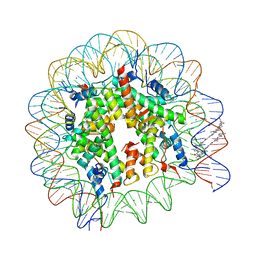 | | LIGAND BINDING ALTERS THE STRUCTURE AND DYNAMICS OF NUCLEOSOMAL DNA | | Descriptor: | 3-AMINO-(DIMETHYLPROPYLAMINE), 4-AMINO-(1-METHYLIMIDAZOLE)-2-CARBOXYLIC ACID, 4-AMINO-(1-METHYLPYRROLE)-2-CARBOXYLIC ACID, ... | | Authors: | Suto, R.K, Edayathumangalam, R.S, White, C.L, Melander, C, Gottesfeld, J.M, Dervan, P.B, Luger, K. | | Deposit date: | 2002-06-18 | | Release date: | 2003-02-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structures of Nucleosome Core Particles in Complex with Minor Groove DNA-binding Ligands
J.MOL.BIOL., 326, 2003
|
|
1M1B
 
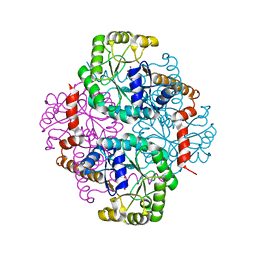 | | Crystal Structure of Phosphoenolpyruvate Mutase Complexed with Sulfopyruvate | | Descriptor: | MAGNESIUM ION, PHOSPHOENOLPYRUVATE PHOSPHOMUTASE, SULFOPYRUVATE | | Authors: | Liu, S, Lu, Z, Jia, Y, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2002-06-18 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Dissociative phosphoryl transfer in PEP mutase catalysis: structure of the enzyme/sulfopyruvate complex and kinetic properties of mutants.
Biochemistry, 41, 2002
|
|
1M1C
 
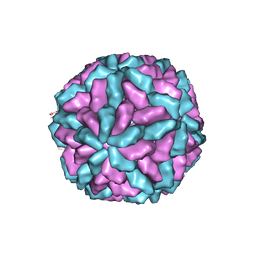 | | Structure of the L-A virus | | Descriptor: | Major coat protein | | Authors: | Naitow, H, Tang, J, Canady, M, Wickner, R.B, Johnson, J.E. | | Deposit date: | 2002-06-18 | | Release date: | 2002-10-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | L-A virus at 3.4 A resolution reveals particle architecture and mRNA decapping mechanism.
Nat.Struct.Biol., 9, 2002
|
|
