1H0Z
 
 | | LEKTI domain six | | Descriptor: | SERINE PROTEASE INHIBITOR KAZAL-TYPE 5, CONTAINS HEMOFILTRATE PEPTIDE HF6478, HEMOFILTRATE PEPTIDE HF7665 | | Authors: | Lauber, T, Roesch, P, Marx, U.C. | | Deposit date: | 2002-07-01 | | Release date: | 2003-06-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Homologous Proteins with Different Folds: The Three-Dimensional Structures of Domains 1 and 6 of the Multiple Kazal-Type Inhibitor Lekti
J.Mol.Biol., 328, 2003
|
|
1H10
 
 | | HIGH RESOLUTION STRUCTURE OF THE PLECKSTRIN HOMOLOGY DOMAIN OF PROTEIN KINASE B/AKT BOUND TO INS(1,3,4,5)-TETRAKISPHOPHATE | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, RAC-ALPHA SERINE/THREONINE KINASE | | Authors: | Thomas, C.C, Deak, M, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2002-07-01 | | Release date: | 2003-06-27 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High Resolution Structure of the Pleckstrin Homology Domain of Protein Kinase B/Akt Bound to Phosphatidylinositol (3,4,5)-Trisphosphate
Curr.Biol., 12, 2002
|
|
1H11
 
 | |
1H12
 
 | | Structure of a cold-adapted family 8 xylanase | | Descriptor: | ENDO-1,4-BETA-XYLANASE, alpha-D-xylopyranose, beta-D-xylopyranose | | Authors: | Van Petegem, F, Collins, T, Meuwis, M.A, Feller, G, Gerday, C, Van Beeumen, J. | | Deposit date: | 2002-07-02 | | Release date: | 2003-03-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Structure of a Cold-Adapted Family 8 Xylanase at 1.3 A Resolution: Structural Adaptations to Cold and Investigation of the Active Site
J.Biol.Chem., 278, 2003
|
|
1H13
 
 | | Structure of a cold-adapted family 8 xylanase | | Descriptor: | ENDO-1,4-BETA-XYLANASE | | Authors: | Van Petegem, F, Collins, T, Meuwis, M.A, Feller, G, Gerday, C, Van Beeumen, J. | | Deposit date: | 2002-07-02 | | Release date: | 2003-03-13 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Structure of a Cold-Adapted Family 8 Xylanase at 1.3 A Resolution: Structural Adaptations to Cold and Investigation of the Active Site
J.Biol.Chem., 278, 2003
|
|
1H14
 
 | | Structure of a cold-adapted family 8 xylanase | | Descriptor: | ENDO-1,4-BETA-XYLANASE | | Authors: | Van Petegem, F, Collins, T, Meuwis, M.A, Feller, G, Gerday, C, Van Beeumen, J. | | Deposit date: | 2002-07-02 | | Release date: | 2003-03-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structure of a Cold-Adapted Family 8 Xylanase at 1.3 A Resolution: Structural Adaptations to Cold and Investigation of the Active Site
J.Biol.Chem., 278, 2003
|
|
1H15
 
 | | X-ray crystal structure of HLA-DRA1*0101/DRB5*0101 complexed with a peptide from Epstein Barr Virus DNA polymerase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA POLYMERASE, ... | | Authors: | Lang, H, Jacobsen, H, Ikemizu, S, Andersson, C, Harlos, K, Madsen, L, Hjorth, P, Sondergaard, L, Svejgaard, A, Wucherpfennig, K, Stuart, D.I, Bell, J.I, Jones, E.Y, Fugger, L. | | Deposit date: | 2002-07-02 | | Release date: | 2002-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A Functional and Structural Basis for Tcr Cross-Reactivity in Multiple Sclerosis
Nat.Immunol., 3, 2002
|
|
1H16
 
 | | Pyruvate Formate-Lyase (E.coli) in complex with Pyruvate and CoA | | Descriptor: | COENZYME A, FORMATE ACETYLTRANSFERASE 1, L-TREITOL, ... | | Authors: | Becker, A, Kabsch, W. | | Deposit date: | 2002-07-03 | | Release date: | 2002-11-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | X-Ray Structure of Pyruvate Formate-Lyase in Complex with Pyruvate and Coa.How the Enzyme Uses the Cys-418 Thiyl Radical for Pyruvate Cleavage
J.Biol.Chem., 277, 2002
|
|
1H17
 
 | |
1H18
 
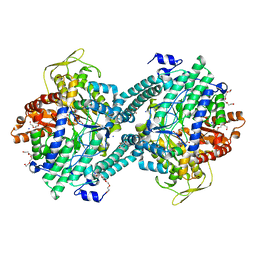 | | Pyruvate Formate-Lyase (E.coli) in complex with Pyruvate | | Descriptor: | FORMATE ACETYLTRANSFERASE 1, L-TREITOL, PYRUVIC ACID, ... | | Authors: | Becker, A, Kabsch, W. | | Deposit date: | 2002-07-04 | | Release date: | 2002-11-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-Ray Structure of Pyruvate Formate-Lyase in Complex with Pyruvate and Coa.How the Enzyme Uses the Cys-418 Thiyl Radical for Pyruvate Cleavage
J.Biol.Chem., 277, 2002
|
|
1H19
 
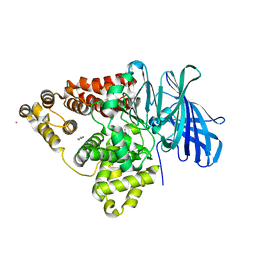 | | STRUCTURE OF [E271Q]LEUKOTRIENE A4 HYDROLASE | | Descriptor: | ACETIC ACID, IMIDAZOLE, LEUKOTRIENE A-4 HYDROLASE, ... | | Authors: | Rudberg, P.C, Tholander, F, Thunnissen, M.M.G.M, Haeggstrom, J.Z. | | Deposit date: | 2002-07-04 | | Release date: | 2002-08-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Leukotriene A4 Hydrolase/Aminopeptidase, Glutamate 271 is a Catalyticresidue with Specific Roles in Two Distinct Enzyme Mechanisms
J.Biol.Chem., 277, 2002
|
|
1H1A
 
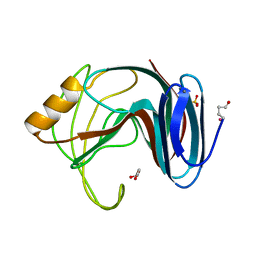 | | Thermophilic beta-1,4-xylanase from Chaetomium thermophilum | | Descriptor: | CALCIUM ION, Endo-1,4-beta-xylanase, GLYCEROL, ... | | Authors: | Hakulinen, N, Rouvinen, J. | | Deposit date: | 2002-07-05 | | Release date: | 2003-07-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Three-Dimensional Structures of Thermophilic Beta-1,4-Xylanases from Chaetomium Thermophilum and Nonomuraea Flexuosa. Comparison of Twelve Xylanases in Relation to Their Thermal Stability.
Eur.J.Biochem., 270, 2003
|
|
1H1B
 
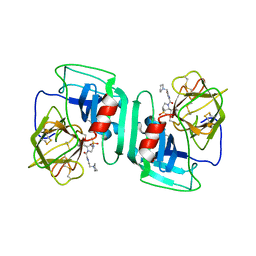 | | Crystal structure of human neutrophil elastase complexed with an inhibitor (GW475151) | | Descriptor: | (2S)-3-METHYL-2-((2R,3S)-3-[(METHYLSULFONYL)AMINO]-1-{[2-(PYRROLIDIN-1-YLMETHYL)-1,3-OXAZOL-4-YL]CARBONYL}PYRROLIDIN-2-YL)BUTANOIC ACID, LEUKOCYTE ELASTASE, alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Macdonald, S.J.F, Dowle, M.D, Harrison, L.A, Clarke, G.D.E, Inglis, G.G.A, Johnson, M.R, Smith, R.A, Amour, A, Fleetwood, G, Humphreys, D.C, Molloy, C.R, Dixon, M, Godward, R.E, Wonacott, A.J, Singh, O.M.P, Hodgson, S.T, Hardy, G.W. | | Deposit date: | 2002-07-05 | | Release date: | 2002-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Further Pyrrolidine Trans-Lactams as Inhibitors of Human Neutrophil Elastase (Hne) with Potential as Development Candidates and the Crystal Structure of Hne Complexed with an Inhibitor (Gw475151)
J.Med.Chem., 45, 2002
|
|
1H1C
 
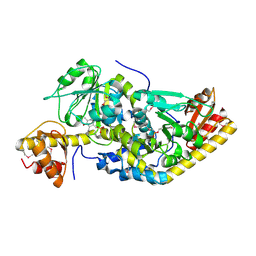 | |
1H1D
 
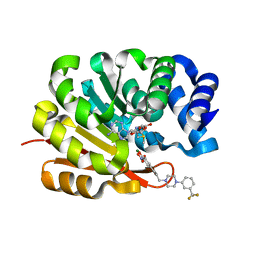 | | Catechol O-Methyltransferase | | Descriptor: | 1-(3,4,DIHYDROXY-5-NITROPHENYL)-3-{4-[3-(TRIFLUOROMETHYL) PHENYL] PIPERAZIN-1-YL}PROPAN-1-ONE, CATECHOL-O-METHYLTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Archer, M, Rodrigues, M.L, Matias, P.M, Bonifacio, M.J, Learmonth, D.A, Soares-da-Silva, P, Carrondo, M.A. | | Deposit date: | 2002-07-12 | | Release date: | 2003-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetics and Crystal Structure of Catechol-O-Methyltransferase Complex with Co-Substrate and a Novel Inhibitor with Potential Therapeutic Application
Mol.Pharmacol., 62, 2002
|
|
1H1H
 
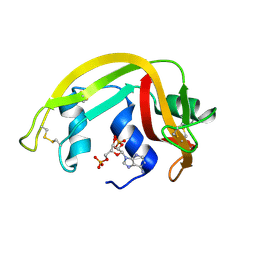 | | Crystal Structure of Eosinophil Cationic Protein in Complex with 2',5'-ADP at 2.0 A resolution Reveals the Details of the Ribonucleolytic Active site | | Descriptor: | ADENOSINE-2'-5'-DIPHOSPHATE, EOSINOPHIL CATIONIC PROTEIN | | Authors: | Mohan, C.G, Boix, E, Evans, H.R, Nikolovski, Z, Nogues, M.V, Cuchillo, C.M, Acharya, K.R. | | Deposit date: | 2002-07-15 | | Release date: | 2002-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Eosinophil Cationic Protein in Complex with 2'5'-Adp at 2.0 A Resolution Reveals the Details of the Ribonucleolytic Active Site
Biochemistry, 41, 2002
|
|
1H1I
 
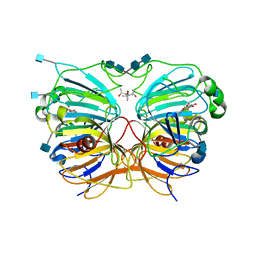 | | CRYSTAL STRUCTURE OF QUERCETIN 2,3-DIOXYGENASE ANAEROBICALLY COMPLEXED WITH THE SUBSTRATE QUERCETN | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Steiner, R.A, Dijkstra, B.W. | | Deposit date: | 2002-07-15 | | Release date: | 2002-11-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Anaerobic Enzyme.Substrate Structures Provide Insight Into the Reaction Mechanism of the Copper- Dependent Quercetin 2,3-Dioxygenase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1H1J
 
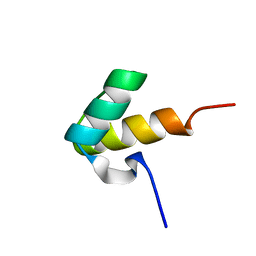 | |
1H1K
 
 | | THE BLUETONGUE VIRUS (BTV) CORE BINDS DSRNA | | Descriptor: | RNA | | Authors: | Diprose, J.M, Grimes, J.M, Sutton, G.C, Burroughs, J.N, Meyer, A, Maan, S, Mertens, P.P.C, Stuart, D.I. | | Deposit date: | 2002-07-17 | | Release date: | 2002-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (10 Å) | | Cite: | The Core of Bluetongue Virus Binds Double-Stranded RNA
J.Virol., 76, 2002
|
|
1H1L
 
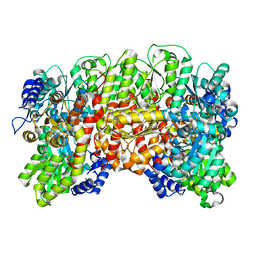 | | NITROGENASE MO-FE PROTEIN FROM KLEBSIELLA PNEUMONIAE, NIFV MUTANT | | Descriptor: | CHLORIDE ION, CITRIC ACID, FE(8)-S(7) CLUSTER, ... | | Authors: | Mayer, S.M, Gormal, C.A, Smith, B.E, Lawson, D.M. | | Deposit date: | 2002-07-18 | | Release date: | 2002-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic Analysis of the Mofe Protein of Nitrogenase from a Nifv Mutant of Klebsiella Pneumoniae Identifies Citrate as a Ligand to the Molybdenum of Iron Molybdenum Cofactor (Femoco).
J.Biol.Chem., 277, 2002
|
|
1H1M
 
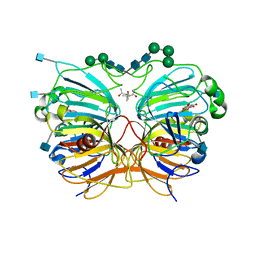 | | CRYSTAL STRUCTURE OF QUERCETIN 2,3-DIOXYGENASE ANAEROBICALLY COMPLEXED WITH THE SUBSTRATE KAEMPFEROL | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Steiner, R.A, Dijkstra, B.W. | | Deposit date: | 2002-07-19 | | Release date: | 2002-11-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Anaerobic Enzyme.Substrate Structures Provide Insight Into the Reaction Mechanism of the Copper- Dependent Quercetin 2,3-Dioxygenase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1H1N
 
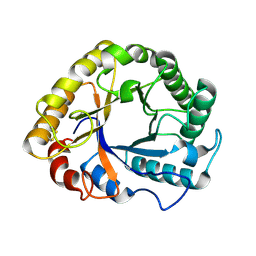 | |
1H1O
 
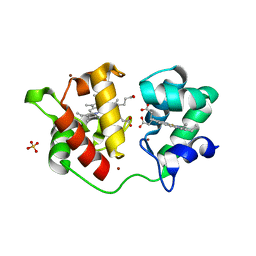 | | Acidithiobacillus ferrooxidans cytochrome c4 structure supports a complex-induced tuning of electron transfer | | Descriptor: | CYTOCHROME C-552, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Abergel, C, Nitschke, W, Malarte, G, Bruschi, M, Claverie, J.-M, Guidici-Orticoni, M.-T. | | Deposit date: | 2002-07-19 | | Release date: | 2003-07-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The Structure of Acidithiobacillus Ferrooxidans C(4)-Cytochrome. A Model for Complex-Induced Electron Transfer Tuning
Structure, 11, 2003
|
|
1H1P
 
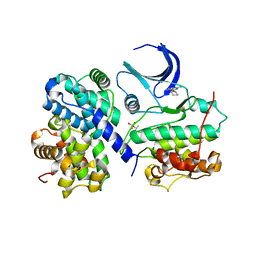 | | Structure of human Thr160-phospho CDK2/cyclin A complexed with the inhibitor NU2058 | | Descriptor: | 6-O-CYCLOHEXYLMETHYL GUANINE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2 | | Authors: | Davies, T.G, Noble, M.E.M, Endicott, J.A, Johnson, L.N. | | Deposit date: | 2002-07-21 | | Release date: | 2002-09-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of a Potent Purine-Based Cyclin-Dependent Kinase Inhibitor
Nat.Struct.Biol., 9, 2002
|
|
1H1Q
 
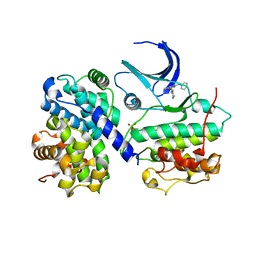 | | Structure of human Thr160-phospho CDK2/cyclin A complexed with the inhibitor NU6094 | | Descriptor: | 2-ANILINO-6-CYCLOHEXYLMETHOXYPURINE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2 | | Authors: | Davies, T.G, Noble, M.E.M, Endicott, J.A, Johnson, L.N. | | Deposit date: | 2002-07-21 | | Release date: | 2002-09-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design of a Potent Purine-Based Cyclin-Dependent Kinase Inhibitor
Nat.Struct.Biol., 9, 2002
|
|
