1ET5
 
 | | CRYSTAL STRUCTURE OF NITRITE REDUCTASE ASP98ASN MUTANT FROM ALCALIGENES FAECALIS S-6 | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE, ZINC ION | | Authors: | Boulanger, M.J, Kukimoto, M, Nishiyama, M, Horinouchi, S, Murphy, M.E.P. | | Deposit date: | 2000-04-12 | | Release date: | 2000-08-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic roles for two water bridged residues (Asp-98 and His-255) in the active site of copper-containing nitrite reductase.
J.Biol.Chem., 275, 2000
|
|
1ET6
 
 | | CRYSTAL STRUCTURE OF THE SUPERANTIGEN SMEZ-2 FROM STREPTOCOCCUS PYOGENES | | Descriptor: | SUPERANTIGEN SMEZ-2 | | Authors: | Arcus, V.L, Proft, T, Sigrell, J.A, Baker, H.M, Fraser, J.D, Baker, E.N. | | Deposit date: | 2000-04-12 | | Release date: | 2000-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conservation and variation in superantigen structure and activity highlighted by the three-dimensional structures of two new superantigens from Streptococcus pyogenes.
J.Mol.Biol., 299, 2000
|
|
1ET7
 
 | | CRYSTAL STRUCTURE OF NITRITE REDUCTASE HIS255ASP MUTANT FROM ALCALIGENES FAECALIS S-6 | | Descriptor: | CADMIUM ION, COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Boulanger, M.J, Kukimoto, M, Nishiyama, M, Horinouchi, S, Murphy, M.E.P. | | Deposit date: | 2000-04-12 | | Release date: | 2000-08-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Catalytic roles for two water bridged residues (Asp-98 and His-255) in the active site of copper-containing nitrite reductase.
J.Biol.Chem., 275, 2000
|
|
1ET8
 
 | | CRYSTAL STRUCTURE OF NITRITE REDUCTASE HIS255ASN MUTANT FROM ALCALIGENES FAECALIS | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE, ZINC ION | | Authors: | Boulanger, M.J, Kukimoto, M, Nishiyama, M, Horinouchi, S, Murphy, M.E.P. | | Deposit date: | 2000-04-12 | | Release date: | 2000-08-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Catalytic roles for two water bridged residues (Asp-98 and His-255) in the active site of copper-containing nitrite reductase.
J.Biol.Chem., 275, 2000
|
|
1ET9
 
 | | CRYSTAL STRUCTURE OF THE SUPERANTIGEN SPE-H FROM STREPTOCOCCUS PYOGENES | | Descriptor: | SUPERANTIGEN SPE-H | | Authors: | Arcus, V.L, Proft, T, Sigrell, J.A, Baker, H.M, Fraser, J.D, Baker, E.N. | | Deposit date: | 2000-04-12 | | Release date: | 2000-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conservation and variation in superantigen structure and activity highlighted by the three-dimensional structures of two new superantigens from Streptococcus pyogenes.
J.Mol.Biol., 299, 2000
|
|
1ETA
 
 | | THE X-RAY CRYSTAL STRUCTURE REFINEMENTS OF NORMAL HUMAN TRANSTHYRETIN AND THE AMYLOIDOGENIC VAL 30-->MET VARIANT TO 1.7 ANGSTROMS RESOLUTION | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, TRANSTHYRETIN | | Authors: | Braden, B.C, Steinrauf, L.K, Hamilton, J.A. | | Deposit date: | 1993-05-12 | | Release date: | 1995-01-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The x-ray crystal structure refinements of normal human transthyretin and the amyloidogenic Val-30-->Met variant to 1.7-A resolution.
J.Biol.Chem., 268, 1993
|
|
1ETB
 
 | | THE X-RAY CRYSTAL STRUCTURE REFINEMENTS OF NORMAL HUMAN TRANSTHYRETIN AND THE AMYLOIDOGENIC VAL 30-->MET VARIANT TO 1.7 ANGSTROMS RESOLUTION | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, TRANSTHYRETIN | | Authors: | Braden, B.C, Steinrauf, L.K, Hamilton, J.A. | | Deposit date: | 1993-05-12 | | Release date: | 1995-01-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The x-ray crystal structure refinements of normal human transthyretin and the amyloidogenic Val-30-->Met variant to 1.7-A resolution.
J.Biol.Chem., 268, 1993
|
|
1ETE
 
 | |
1ETF
 
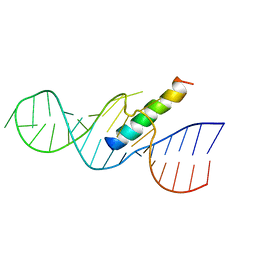 | | REV RESPONSE ELEMENT (RRE) RNA COMPLEXED WITH REV PEPTIDE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | REV PEPTIDE, REV RESPONSE ELEMENT RNA | | Authors: | Battiste, J.L, Mao, H, Rao, N.S, Tan, R, Muhandiram, D.R, Kay, L.E, Frankel, A.D, Willamson, J.R. | | Deposit date: | 1996-08-28 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Alpha helix-RNA major groove recognition in an HIV-1 rev peptide-RRE RNA complex.
Science, 273, 1996
|
|
1ETG
 
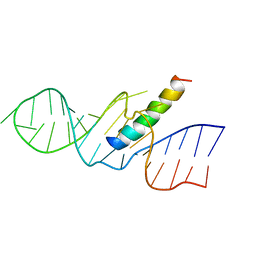 | | REV RESPONSE ELEMENT (RRE) RNA COMPLEXED WITH REV PEPTIDE, NMR, 19 STRUCTURES | | Descriptor: | REV PEPTIDE, REV RESPONSIVE ELEMENT RNA | | Authors: | Battiste, J.L, Mao, H, Rao, N.S, Tan, R, Muhandiram, D.R, Kay, L.E, Frankel, A.D, Willamson, J.R. | | Deposit date: | 1996-08-28 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Alpha helix-RNA major groove recognition in an HIV-1 rev peptide-RRE RNA complex.
Science, 273, 1996
|
|
1ETH
 
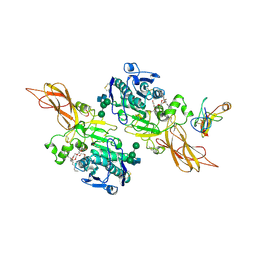 | | TRIACYLGLYCEROL LIPASE/COLIPASE COMPLEX | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, BETA-MERCAPTOETHANOL, CALCIUM ION, ... | | Authors: | Hermoso, J, Pignol, D, Kerfelec, B, Crenon, I, Chapus, C, Fontecilla-Camps, J.C. | | Deposit date: | 1995-09-13 | | Release date: | 1996-12-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Lipase activation by nonionic detergents. The crystal structure of the porcine lipase-colipase-tetraethylene glycol monooctyl ether complex.
J.Biol.Chem., 271, 1996
|
|
1ETJ
 
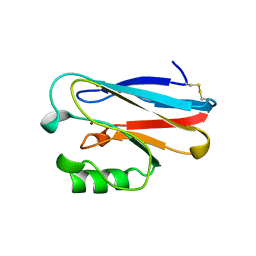 | | AZURIN MUTANT WITH MET 121 REPLACED BY GLU | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Karlsson, B.G, Tsai, L.-C, Nar, H, Sanders-Loehr, J, Bonander, N, Langer, V, Sjolin, L. | | Deposit date: | 1997-01-11 | | Release date: | 1997-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structure determination and characterization of the Pseudomonas aeruginosa azurin mutant Met121Glu.
Biochemistry, 36, 1997
|
|
1ETK
 
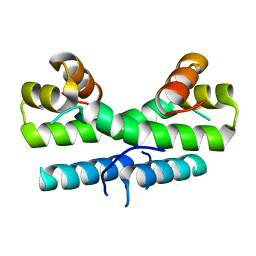 | |
1ETL
 
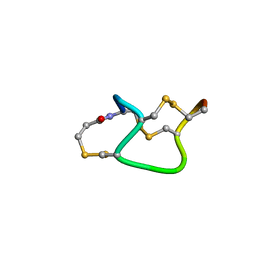 | |
1ETM
 
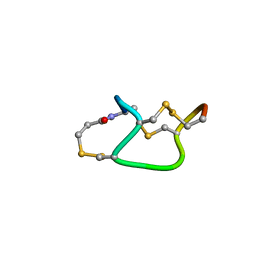 | |
1ETN
 
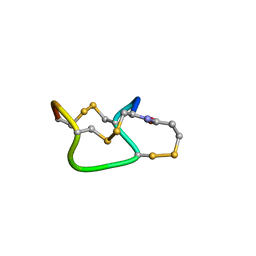 | |
1ETO
 
 | |
1ETP
 
 | |
1ETQ
 
 | |
1ETR
 
 | | REFINED 2.3 ANGSTROMS X-RAY CRYSTAL STRUCTURE OF BOVINE THROMBIN COMPLEXES FORMED WITH THE BENZAMIDINE AND ARGININE-BASED THROMBIN INHIBITORS NAPAP, 4-TAPAP AND MQPA: A STARTING POINT FOR IMPROVING ANTITHROMBOTICS | | Descriptor: | EPSILON-THROMBIN, amino{[(4S)-5-[(2R,4R)-2-carboxy-4-methylpiperidin-1-yl]-4-({[(3R)-3-methyl-1,2,3,4-tetrahydroquinolin-8-yl]sulfonyl}amino)-5-oxopentyl]amino}methaniminium | | Authors: | Bode, W, Brandstetter, H. | | Deposit date: | 1992-07-06 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined 2.3 A X-ray crystal structure of bovine thrombin complexes formed with the benzamidine and arginine-based thrombin inhibitors NAPAP, 4-TAPAP and MQPA. A starting point for improving antithrombotics.
J.Mol.Biol., 226, 1992
|
|
1ETS
 
 | |
1ETT
 
 | |
1ETU
 
 | | STRUCTURAL DETAILS OF THE BINDING OF GUANOSINE DIPHOSPHATE TO ELONGATION FACTOR TU FROM E. COLI AS STUDIED BY X-RAY CRYSTALLOGRAPHY | | Descriptor: | ELONGATION FACTOR TU, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Clark, B.F.C, Lacour, T.F.M, Kjeldgaard, M, Morikawa, K, Nyborg, J, Rubin, R, Thirup, S. | | Deposit date: | 1988-01-15 | | Release date: | 1988-07-16 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural details of the binding of guanosine diphosphate to elongation factor Tu from E. coli as studied by X-ray crystallography.
EMBO J., 4, 1985
|
|
1ETV
 
 | |
1ETW
 
 | |
