2LV8
 
 | | Solution NMR Structure de novo designed rossmann 2x2 fold protein, Northeast Structural Genomics Consortium (NESG) Target OR16 | | 分子名称: | De novo designed rossmann 2x2 fold protein | | 著者 | Liu, G, Koga, R, Koga, N, Xiao, R, Pederson, K, Hamilton, K, Ciccosanti, C, Acton, T.B, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2012-06-29 | | 公開日 | 2012-08-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Principles for designing ideal protein structures.
Nature, 491, 2012
|
|
5Z9J
 
 | | Identification of the functions of unusual cytochrome p450-like monooxygenases involved in microbial secondary metablism | | 分子名称: | PROTOPORPHYRIN IX CONTAINING FE, Putative P450-like enzyme, TRIS(HYDROXYETHYL)AMINOMETHANE | | 著者 | Lu, M, Lin, L, Zhang, C, Chen, Y. | | 登録日 | 2018-02-03 | | 公開日 | 2019-02-06 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Riboflavin Is Directly Involved in the N-Dealkylation Catalyzed by Bacterial Cytochrome P450 Monooxygenases.
Chembiochem, 2020
|
|
1EGL
 
 | |
1EXR
 
 | |
1EYP
 
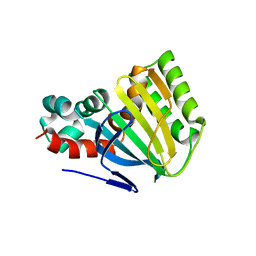 | | CHALCONE ISOMERASE | | 分子名称: | CHALCONE-FLAVONONE ISOMERASE 1 | | 著者 | Jez, J.M, Bowman, M.E, Dixon, R.A, Noel, J.P. | | 登録日 | 2000-05-08 | | 公開日 | 2000-09-06 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure and mechanism of the evolutionarily unique plant enzyme chalcone isomerase.
Nat.Struct.Biol., 7, 2000
|
|
1TIZ
 
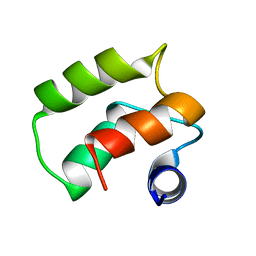 | | Solution Structure of a Calmodulin-Like Calcium-Binding Domain from Arabidopsis thaliana | | 分子名称: | calmodulin-related protein, putative | | 著者 | Song, J, Zhao, Q, Thao, S, Frederick, R.O, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2004-06-02 | | 公開日 | 2004-08-10 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Letter to the Editor: Solution structure of a calmodulin-like calcium-binding domain from Arabidopsis thaliana
J.Biomol.NMR, 30, 2004
|
|
1EGT
 
 | |
1ACW
 
 | | SOLUTION NMR STRUCTURE OF P01, A NATURAL SCORPION PEPTIDE STRUCTURALLY ANALOGOUS TO SCORPION TOXINS SPECIFIC FOR APAMIN-SENSITIVE POTASSIUM CHANNEL, 25 STRUCTURES | | 分子名称: | NATURAL SCORPION PEPTIDE P01 | | 著者 | Blanc, E, Fremont, V, Sizun, P, Meunier, S, Van Rietschoten, J, Thevand, A, Bernassau, J.M, Darbon, H. | | 登録日 | 1997-02-10 | | 公開日 | 1997-04-01 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of P01, a natural scorpion peptide structurally analogous to scorpion toxins specific for apamin-sensitive potassium channel.
Proteins, 24, 1996
|
|
5Z9I
 
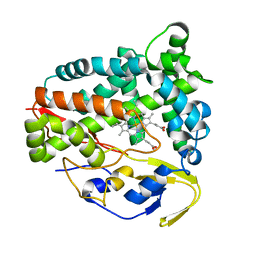 | |
1E9I
 
 | | Enolase from E.coli | | 分子名称: | ENOLASE, MAGNESIUM ION, SULFATE ION | | 著者 | Kuhnel, K, Carpousis, A.J, Luisi, B. | | 登録日 | 2000-10-17 | | 公開日 | 2001-03-15 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Crystal Structure of the Escherichia Coli RNA Degradosome Component Enolase
J.Mol.Biol., 313, 2001
|
|
1A2Q
 
 | | SUBTILISIN BPN' MUTANT 7186 | | 分子名称: | ACETONE, CALCIUM ION, SUBTILISIN BPN' | | 著者 | Gilliland, G.L, Whitlow, M, Howard, A.J. | | 登録日 | 1998-01-08 | | 公開日 | 1998-04-29 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Large increases in general stability for subtilisin BPN' through incremental changes in the free energy of unfolding.
Biochemistry, 28, 1989
|
|
1EAO
 
 | | THE RUNX1 Runt domain at 1.4A resolution: a structural switch and specifically bound chloride ions modulate DNA binding | | 分子名称: | BROMIDE ION, RUNT-RELATED TRANSCRIPTION FACTOR 1 | | 著者 | Backstrom, S, Wolf-Watz, M, Grundstrom, C, Hard, T.H, Grundstrom, T, Sauer, U.H. | | 登録日 | 2001-07-14 | | 公開日 | 2002-09-12 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | The Runx1 Runt Domain at 1.25A Resolution: A Structural Switch and Specifically Bound Chloride Ions Modulate DNA Binding
J.Mol.Biol., 322, 2002
|
|
1E5W
 
 | |
1F0W
 
 | |
1EZM
 
 | |
1ALE
 
 | |
1JSA
 
 | | MYRISTOYLATED RECOVERIN WITH TWO CALCIUMS BOUND, NMR, 24 STRUCTURES | | 分子名称: | CALCIUM ION, MYRISTIC ACID, RECOVERIN | | 著者 | Ames, J.B, Ishima, R, Tanaka, T, Gordon, J.I, Stryer, L, Ikura, M. | | 登録日 | 1997-06-04 | | 公開日 | 1997-10-15 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Molecular mechanics of calcium-myristoyl switches.
Nature, 389, 1997
|
|
5PAH
 
 | |
186D
 
 | |
1A93
 
 | | NMR SOLUTION STRUCTURE OF THE C-MYC-MAX HETERODIMERIC LEUCINE ZIPPER, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | MAX PROTEIN, MYC PROTO-ONCOGENE PROTEIN | | 著者 | Lavigne, P, Crump, M.P, Gagne, S.M, Hodges, R.S, Kay, C.M, Sykes, B.D. | | 登録日 | 1998-04-15 | | 公開日 | 1998-10-21 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Insights into the mechanism of heterodimerization from the 1H-NMR solution structure of the c-Myc-Max heterodimeric leucine zipper.
J.Mol.Biol., 281, 1998
|
|
5OQS
 
 | | Solution structure of antifungal protein NFAP | | 分子名称: | NFAP | | 著者 | Hajdu, D, Czajlik, A, Marx, F, Galgoczy, L, Batta, G. | | 登録日 | 2017-08-14 | | 公開日 | 2018-07-25 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and novel insights into phylogeny and mode of action of the Neosartorya (Aspergillus) fischeri antifungal protein (NFAP).
Int.J.Biol.Macromol., 129, 2019
|
|
1AAJ
 
 | |
1F3P
 
 | | FERREDOXIN REDUCTASE (BPHA4)-NADH COMPLEX | | 分子名称: | FERREDOXIN REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Senda, T, Yamada, T, Sakurai, N, Kubota, M, Nishizaki, T, Masai, E, Fukuda, M, Mitsuidagger, Y. | | 登録日 | 2000-06-06 | | 公開日 | 2001-06-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of NADH-dependent ferredoxin reductase component in biphenyl dioxygenase.
J.Mol.Biol., 304, 2000
|
|
1X93
 
 | | NMR Structure of Helicobacter pylori HP0222 | | 分子名称: | hypothetical protein HP0222 | | 著者 | Popescu, A, Karpay, A, Israel, D, Peek Jr, R.M, Krezel, A.M. | | 登録日 | 2004-08-19 | | 公開日 | 2005-03-22 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Helicobacter pylori protein HP0222 belongs to Arc/MetJ family of transcriptional regulators.
Proteins, 59, 2005
|
|
1F53
 
 | | NMR STRUCTURE OF KILLER TOXIN-LIKE PROTEIN SKLP | | 分子名称: | YEAST KILLER TOXIN-LIKE PROTEIN | | 著者 | Ohki, S, Kariya, E, Hiraga, K, Wakamiya, A, Isobe, T, Oda, K, Kainosho, M. | | 登録日 | 2000-06-12 | | 公開日 | 2000-12-27 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of Streptomyces killer toxin-like protein, SKLP: further evidence for the wide distribution of single-domain betagamma-crystallin superfamily proteins.
J.Mol.Biol., 305, 2001
|
|
