4UVV
 
 | | Crystal structure of human tankyrase 2 in complex with 3-(4- chlorophenyl)-5-methyl-1,2-dihydroisoquinolin-1-one | | 分子名称: | 3-(4-chlorophenyl)-5-methylisoquinolin-1(2H)-one, GLYCEROL, SULFATE ION, ... | | 著者 | Haikarainen, T, Narwal, M, Lehtio, L. | | 登録日 | 2014-08-08 | | 公開日 | 2015-07-29 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Exploration of the Nicotinamide-Binding Site of the Tankyrases, Identifying 3-Arylisoquinolin-1-Ones as Potent and Selective Inhibitors in Vitro.
Bioorg.Med.Chem., 23, 2015
|
|
4V1X
 
 | | The structure of the hexameric atrazine chlorohydrolase, AtzA | | 分子名称: | ATRAZINE CHLOROHYDROLASE, DI(HYDROXYETHYL)ETHER, FE (III) ION | | 著者 | Peat, T.S, Newman, J, Balotra, S, Lucent, D, Warden, A.C, Scott, C. | | 登録日 | 2014-10-04 | | 公開日 | 2015-03-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The Structure of the Hexameric Atrazine Chlorohydrolase Atza.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UZX
 
 | | High-resolution NMR structures of the domains of Saccharomyces cerevisiae Tho1 | | 分子名称: | PROTEIN THO1 | | 著者 | Jacobsen, J.O.B, Allen, M.D, Freund, S.M.V, Bycroft, M. | | 登録日 | 2014-09-09 | | 公開日 | 2014-12-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | High-Resolution NMR Structures of the Domains of Saccharomyces Cerevisiae Tho1.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4UZM
 
 | |
4UU9
 
 | | Crystal structure of the human c5a in complex with MEDI7814 a neutralising antibody | | 分子名称: | COMPLEMENT C5, MEDI7814, SULFATE ION | | 著者 | Colley, C, Sridharan, S, Dobson, C, Popovic, B, Debreczeni, J.E, Hargreaves, D, Edwards, B, Brennan, J, England, L, Fung, S, An Eghobamien, L, Sivars, U, Woods, R, Flavell, L, Renshaw, G.J, Wickson, K, Wilkinson, T, Davies, R, Bonnell, J, Warrener, P, Howes, R, Vaughan, T. | | 登録日 | 2014-07-25 | | 公開日 | 2015-08-12 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Structure and characterization of a high affinity C5a monoclonal antibody that blocks binding to C5aR1 and C5aR2 receptors.
MAbs, 10, 2018
|
|
4UZ0
 
 | | Crystal Structure of apoptosis repressor with CARD (ARC) | | 分子名称: | GLYCEROL, NUCLEOLAR PROTEIN 3 | | 著者 | Kim, S.H, Jeong, J.H, Jang, T.H, Kim, Y.G, Park, H.H. | | 登録日 | 2014-09-04 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.399 Å) | | 主引用文献 | Crystal Structure of Caspase Recruiting Domain (Card) of Apoptosis Repressor with Card (Arc) and its Implication in Inhibition of Apoptosis.
Sci.Rep., 5, 2015
|
|
4V1K
 
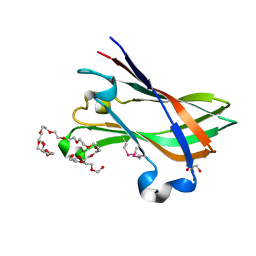 | | SeMet structure of a novel carbohydrate binding module from glycoside hydrolase family 9 (Cel9A) from Ruminococcus flavefaciens FD-1 | | 分子名称: | 2-HYDROXY BUTANE-1,4-DIOL, CALCIUM ION, CARBOHYDRATE BINDING MODULE, ... | | 著者 | Venditto, I, Goyal, A, Thompson, A, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | 登録日 | 2014-09-29 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Complexity of the Ruminococcus Flavefaciens Cellulosome Reflects an Expansion in Glycan Recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4V34
 
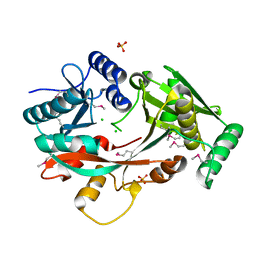 | | The Structure of A-PGS from Pseudomonas aeruginosa (SeMet derivative) | | 分子名称: | ALANYL-TRNA-DEPENDENT L-ALANYL- PHOPHATIDYLGLYCEROL SYNTHASE, CHLORIDE ION, SULFATE ION | | 著者 | Krausze, J, Hebecker, S, Hasenkampf, T, Heinz, D.W, Moser, J. | | 登録日 | 2014-10-16 | | 公開日 | 2015-08-19 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structures of Two Bacterial Resistance Factors Mediating tRNA-Dependent Aminoacylation of Phosphatidylglycerol with Lysine or Alanine.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3S4T
 
 | | Crystal structure of putative amidohydrolase-2 (EFI-target 500288)from Polaromonas sp. JS666 | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, Amidohydrolase 2, ... | | 著者 | Ramagopal, U.A, Toro, R, Girlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2011-05-20 | | 公開日 | 2011-08-24 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of putative amidohydrolase-2 (EFI-target 500288)from Polaromonas sp. JS666
To be published
|
|
4UWW
 
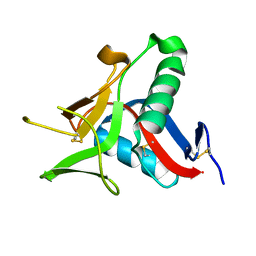 | | Crystallographic Structure of the Intramineral Protein Struthicalcin from Struthio camelus Eggshell | | 分子名称: | STRUTHIOCALCIN-1 | | 著者 | Ruiz, R.R, Moreno, A, Romero, A. | | 登録日 | 2014-08-14 | | 公開日 | 2015-04-08 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | Crystal Structure of Struthiocalcin-1, an Intramineral Protein from Struthio Camelus Eggshell, in Two Different Crystal Forms.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3K6A
 
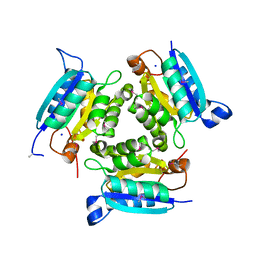 | |
3S75
 
 | |
4V2D
 
 | | FLRT2 LRR domain | | 分子名称: | FIBRONECTIN LEUCINE RICH TRANSMEMBRANE PROTEIN 2 | | 著者 | Seiradake, E, del Toro, D, Nagel, D, Cop, F, Haertl, R, Ruff, T, Seyit-Bremer, G, Harlos, K, Border, E.C, Acker-Palmer, A, Jones, E.Y, Klein, R. | | 登録日 | 2014-10-08 | | 公開日 | 2014-11-05 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Flrt Structure: Balancing Repulsion and Cell Adhesion in Cortical and Vascular Development
Neuron, 84, 2014
|
|
4V0G
 
 | | JAK3 in complex with a covalent EGFR inhibitor | | 分子名称: | N-[3-(2-{3-amino-6-[1-(1-methylpiperidin-4-yl)-1H-pyrazol-4-yl]pyrazin-2-yl}-1H-benzimidazol-1-yl)phenyl]propanamide, TYROSINE-PROTEIN KINASE JAK3 | | 著者 | Debreczeni, J.E, Hennessy, E.J, Chuaquini, C, Ashton, S, Coclough, N, Cross, D.A.E, Eberlein, C, Gingipalli, L, Klinowska, T.C.M, Orme, J.P, Sha, L, Wu, X. | | 登録日 | 2014-09-16 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Utilisation of Structure Based Design to Identify Novel, Irreversible Inhibitors of the Epidermal Growth Factor Receptor (Egfr) Harboring the Gatekeeper T790M Mutation
To be Published
|
|
4V1R
 
 | | Structure of a selenomethionine derivative of the GH76 alpha- mannanase BT2949 Bacteroides thetaiotaomicron | | 分子名称: | 1,2-ETHANEDIOL, ALPHA-1,6-MANNANASE, S,R MESO-TARTARIC ACID | | 著者 | Thompson, A.J, Cuskin, F, Spears, R.J, Dabin, J, Turkenburg, J.P, Gilbert, H.J, Davies, G.J. | | 登録日 | 2014-10-02 | | 公開日 | 2015-02-11 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of the Gh76 Alpha-Mannanase Homolog, Bt2949, from the Gut Symbiont Bacteroides Thetaiotaomicron
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UW7
 
 | | Structure of the carboxy-terminal domain of the bacteriophage T5 L- shaped tail fiber without its intra-molecular chaperone domain | | 分子名称: | GLYCEROL, L-SHAPED TAIL FIBER PROTEIN | | 著者 | Garcia-Doval, C, Luque, D, Caston, J.R, Otero, J.M, Llamas-Saiz, A.L, Boulanger, P, van Raaij, M.J. | | 登録日 | 2014-08-08 | | 公開日 | 2015-08-05 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | Structure of the Receptor-Binding Carboxy-Terminal Domain of the Bacteriophage T5 L-Shaped Tail Fibre with and without Its Intra-Molecular Chaperone.
Viruses, 7, 2015
|
|
3S8F
 
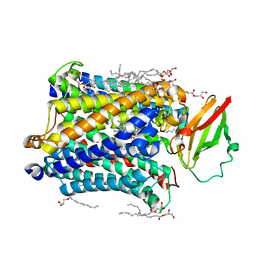 | | 1.8 A structure of ba3 cytochrome c oxidase from Thermus thermophilus in lipid environment | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, ... | | 著者 | Tiefenbrunn, T, Liu, W, Chen, Y, Katritch, V, Stout, C.D, Fee, J.A, Cherezov, V. | | 登録日 | 2011-05-27 | | 公開日 | 2011-08-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | High resolution structure of the ba3 cytochrome c oxidase from Thermus thermophilus in a lipidic environment.
Plos One, 6, 2011
|
|
4V2B
 
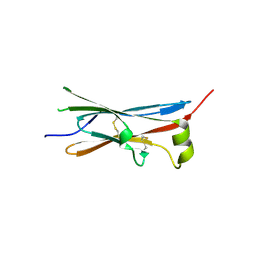 | | rat Unc5D Ig domain 1 | | 分子名称: | PROTEIN UNC5D | | 著者 | Seiradake, E, del Toro, D, Nagel, D, Cop, F, Haertl, R, Ruff, T, Seyit-Bremer, G, Harlos, K, Border, E.C, Acker-Palmer, A, Jones, E.Y, Klein, R. | | 登録日 | 2014-10-08 | | 公開日 | 2014-11-05 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Flrt Structure: Balancing Repulsion and Cell Adhesion in Cortical and Vascular Development
Neuron, 84, 2014
|
|
4USE
 
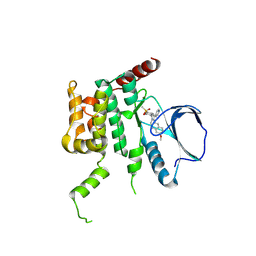 | | Human STK10 (LOK) with SB-633825 | | 分子名称: | 4-{5-(6-methoxynaphthalen-2-yl)-1-methyl-2-[2-methyl-4-(methylsulfonyl)phenyl]-1H-imidazol-4-yl}pyridine, SERINE/THREONINE-PROTEIN KINASE 10 | | 著者 | Elkins, J.M, Salah, E, Szklarz, M, von Delft, F, Canning, P, Raynor, J, Bountra, C, Edwards, A.M, Knapp, S. | | 登録日 | 2014-07-07 | | 公開日 | 2015-07-22 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Comprehensive Characterization of the Published Kinase Inhibitor Set.
Nat.Biotechnol., 34, 2016
|
|
3SH3
 
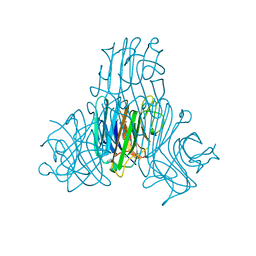 | | Crystal structure of a pro-inflammatory lectin from the seeds of Dioclea wilsonii STANDL | | 分子名称: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Rangel, T.B.A, Rocha, B.A.M, Bezerra, G.A, Bezerra, M.J.B, Nascimento, K.S, Nagano, C.S, Sampaio, A.H, Assreuy, A.M.S, Gruber, K, Delatorre, P, Cavada, B.S. | | 登録日 | 2011-06-15 | | 公開日 | 2011-10-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of a pro-inflammatory lectin from the seeds of Dioclea wilsonii Standl.
Biochimie, 94, 2012
|
|
4US7
 
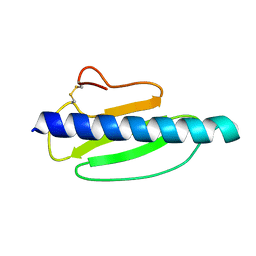 | | Sulfur SAD Phased Structure of a Type IV Pilus Protein from Shewanella oneidensis | | 分子名称: | PILD PROCESSED PROTEIN, SODIUM ION, SULFATE ION | | 著者 | Gorgel, M, Boeggild, A, Ulstrup, J.J, Mueller, U, Weiss, M, Nissen, P, Boesen, T. | | 登録日 | 2014-07-03 | | 公開日 | 2015-04-29 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | High-Resolution Structure of a Type Iv Pilin from the Metal- Reducing Bacterium Shewanella Oneidensis.
Bmc Struct.Biol., 15, 2015
|
|
3SHF
 
 | |
4UW9
 
 | |
4UXE
 
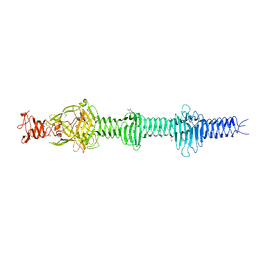 | | Crystal structure of the carboxy-terminal region of the bacteriophage T4 proximal long tail fibre protein gp34, P21 selenomethionine crystal | | 分子名称: | GLYCEROL, LARGE TAIL FIBER PROTEIN P34 | | 著者 | Granell, M, Alvira, S, Garcia-Doval, C, Singh, A.K, van Raaij, M.J. | | 登録日 | 2014-08-22 | | 公開日 | 2015-08-19 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of the Carboxy-Terminal Region of the Bacteriophage T4 Proximal Long Tail Fiber Protein Gp34.
Viruses, 9, 2017
|
|
3SA1
 
 | |
