3L6I
 
 | | Crystal structure of the uncharacterized lipoprotein yceb from e. coli at the resolution 2.0a. northeast structural genomics consortium target er542 | | 分子名称: | SODIUM ION, Uncharacterized lipoprotein yceB | | 著者 | Kuzin, A.P, Neely, H, Seetharaman, J, Chen, C.X, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2009-12-23 | | 公開日 | 2010-01-26 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.011 Å) | | 主引用文献 | Crystal structure of the uncharacterized lipoprotein yceb from e. coli at the resolution 2.0a. northeast structural genomics consortium target er542
To be Published
|
|
3L70
 
 | | Cytochrome BC1 complex from chicken with trifloxystrobin bound | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CARDIOLIPIN, Coenzyme Q10, ... | | 著者 | Huang, L, Berry, E.A. | | 登録日 | 2009-12-27 | | 公開日 | 2010-02-02 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Famoxadone and related inhibitors bind like methoxy acrylate inhibitors in the Qo site of the BC1 compl and fix the rieske iron-sulfur protein in a positio close to but distinct from that seen with stigmatellin and other "distal" Qo inhibitors.
To be Published
|
|
7NKR
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8210 | | 分子名称: | Nb8210, Polymerase acidic protein, Polymerase basic protein 2,Polymerase basic protein 2, ... | | 著者 | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | 登録日 | 2021-02-18 | | 公開日 | 2021-12-01 | | 最終更新日 | 2022-02-09 | | 実験手法 | ELECTRON MICROSCOPY (5.6 Å) | | 主引用文献 | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
5H61
 
 | |
5H6I
 
 | | Crystal Structure of GBS CAMP Factor | | 分子名称: | CHLORIDE ION, Protein B, SULFATE ION | | 著者 | Jin, T.C, Brefo-Mensah, E.K. | | 登録日 | 2016-11-13 | | 公開日 | 2017-11-22 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Crystal structure of theStreptococcus agalactiaeCAMP factor provides insights into its membrane-permeabilizing activity.
J.Biol.Chem., 293, 2018
|
|
3L75
 
 | | Cytochrome BC1 complex from chicken with fenamidone bound | | 分子名称: | (5S)-5-methyl-2-(methylsulfanyl)-5-phenyl-3-(phenylamino)-3,5-dihydro-4H-imidazol-4-one, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, AZIDE ION, ... | | 著者 | Huang, L, Berry, E.A. | | 登録日 | 2009-12-28 | | 公開日 | 2010-02-02 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | FAMOXADONE AND RELATED INHIBITORS BIND LIKE METHOXY ACRYLATE INHIBITORS IN THE Qo SITE OF THE BC1 COMPL AND FIX THE RIESKE IRON-SULFUR PROTEIN IN A POSITIO CLOSE TO BUT DISTINCT FROM THAT SEEN WITH STIGMATELLIN AND OTHER "DISTAL" Qo INHIBITORS.
To be Published
|
|
6OBE
 
 | | Ricin A chain bound to VHH antibody V6H8 | | 分子名称: | 1,2-ETHANEDIOL, IMIDAZOLE, NICKEL (II) ION, ... | | 著者 | Rudolph, M.J. | | 登録日 | 2019-03-20 | | 公開日 | 2020-04-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.732 Å) | | 主引用文献 | Intracellular Neutralization of Ricin Toxin by Single-domain Antibodies Targeting the Active Site.
J.Mol.Biol., 432, 2020
|
|
5H6S
 
 | | Crystal structure of Hydrazidase S179A mutant complexed with a substrate | | 分子名称: | 4-oxidanylbenzohydrazide, Amidase | | 著者 | Akiyama, T, Ishii, M, Takuwa, A, Oinuma, K, Sasaki, Y, Takaya, N, Yajima, S. | | 登録日 | 2016-11-15 | | 公開日 | 2017-02-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis of the substrate recognition of hydrazidase isolated from Microbacterium sp. strain HM58-2, which catalyzes acylhydrazide compounds as its sole carbon source
Biochem. Biophys. Res. Commun., 482, 2017
|
|
3L84
 
 | | High resolution crystal structure of transketolase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | 分子名称: | ACETATE ION, GLYCEROL, Transketolase | | 著者 | Nocek, B, Makowska-Grzyska, M, Maltseva, N, Grimshaw, S, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2009-12-29 | | 公開日 | 2010-02-09 | | 最終更新日 | 2012-02-22 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | High resolution crystal structure of transketolase from Campylobacter jejuni subsp. jejuni NCTC 11168
To be Published
|
|
5H7H
 
 | | Crystal structure of the BCL6 BTB domain in complex with F1324(10-13) | | 分子名称: | 1,2-ETHANEDIOL, B-cell lymphoma 6 protein, F1324 peptide residues 10-13 | | 著者 | Sogabe, S, Ida, K, Lane, W, Snell, G. | | 登録日 | 2016-11-18 | | 公開日 | 2016-12-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Discovery of high-affinity BCL6-binding peptide and its structure-activity relationship.
Biochem. Biophys. Res. Commun., 482, 2017
|
|
5H8P
 
 | |
5H94
 
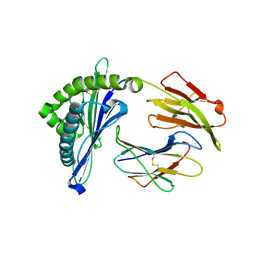 | | Crystal structure of Swine MHC CLASSI for 1.48 angstroms | | 分子名称: | Beta-2-microglobulin, MHC class I antigen, Nonapeptide from Influenza A virus HA protein | | 著者 | Fan, S, Zhang, N, Wang, S, Wu, Y, Xia, C. | | 登録日 | 2015-12-25 | | 公開日 | 2016-05-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Structural and Biochemical Analyses of Swine Major Histocompatibility Complex Class I Complexes and Prediction of the Epitope Map of Important Influenza A Virus Strains
J.Virol., 90, 2016
|
|
3L8I
 
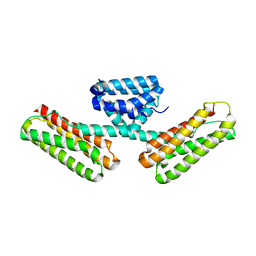 | | Crystal structure of CCM3, a cerebral cavernous malformation protein critical for vascular integrity | | 分子名称: | Programmed cell death protein 10 | | 著者 | Li, X, Zhang, R, Zhang, H, He, Y, Ji, W, Min, W, Boggon, T.J. | | 登録日 | 2009-12-31 | | 公開日 | 2010-05-19 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of CCM3, a cerebral cavernous malformation protein critical for vascular integrity.
J.Biol.Chem., 285, 2010
|
|
3LAQ
 
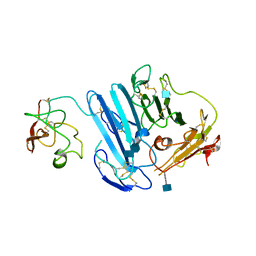 | | Structure-based engineering of species selectivity in the uPA-uPAR interaction | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Urokinase plasminogen activator surface receptor, Urokinase-type plasminogen activator | | 著者 | Huang, M. | | 登録日 | 2010-01-06 | | 公開日 | 2010-02-02 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structure-based engineering of species selectivity in the interaction between urokinase and its receptor: implication for preclinical cancer therapy.
J.Biol.Chem., 285, 2010
|
|
5HA1
 
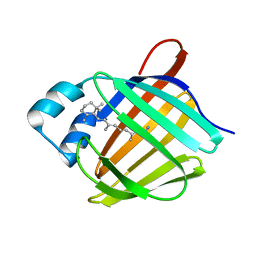 | | Crystal structure of human cellular retinol binding protein 1 in complex with retinylamine | | 分子名称: | (2~{E},4~{E},6~{E},8~{E})-3,7-dimethyl-9-(2,6,6-trimethylcyclohexen-1-yl)nona-2,4,6,8-tetraen-1-amine, Retinol-binding protein 1 | | 著者 | Golczak, M, Arne, J.M, Silvaroli, J.A, Kiser, P.D, Banerjee, S. | | 登録日 | 2015-12-29 | | 公開日 | 2016-03-02 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Ligand Binding Induces Conformational Changes in Human Cellular Retinol-binding Protein 1 (CRBP1) Revealed by Atomic Resolution Crystal Structures.
J.Biol.Chem., 291, 2016
|
|
7NK2
 
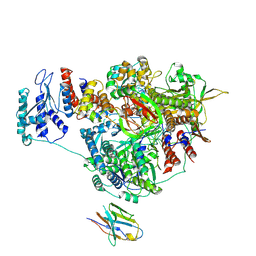 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8202 core | | 分子名称: | Nanobody8202, Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, ... | | 著者 | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | 登録日 | 2021-02-17 | | 公開日 | 2021-12-01 | | 最終更新日 | 2022-02-09 | | 実験手法 | ELECTRON MICROSCOPY (4.84 Å) | | 主引用文献 | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
3LB2
 
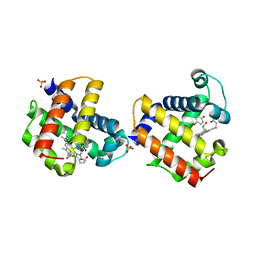 | | Two-site competitive inhibition in dehaloperoxidase-hemoglobin | | 分子名称: | 4-BROMOPHENOL, DEHALOPEROXIDASE A, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | de Serrano, V.S, Franzen, S, Thompson, M.K, Davis, M.F, Nicoletti, F.P, Howes, B.D, Smulevich, G. | | 登録日 | 2010-01-07 | | 公開日 | 2010-11-24 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.06 Å) | | 主引用文献 | Internal binding of halogenated phenols in dehaloperoxidase-hemoglobin inhibits peroxidase function.
Biophys.J., 99, 2010
|
|
5HA9
 
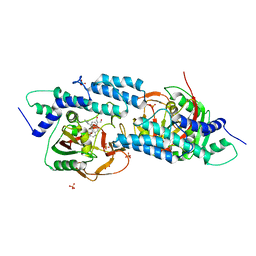 | | Crystal structure-based design and disovery of a novel PARP1 antiagonist (BL-PA10) that induces apoptosis and inhibits metastasis in triple negative breast cancer | | 分子名称: | Amitriptyline, GLYCEROL, Poly [ADP-ribose] polymerase 1, ... | | 著者 | Fu, L, Peng, H, Zhang, L, Ouyang, L. | | 登録日 | 2015-12-30 | | 公開日 | 2016-03-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (4.01 Å) | | 主引用文献 | Crystal structure-based discovery of a novel synthesized PARP1 inhibitor (OL-1) with apoptosis-inducing mechanisms in triple-negative breast cancer.
Sci Rep, 6, 2016
|
|
5HAM
 
 | |
3L9I
 
 | | Myosin VI nucleotide-free (mdinsert2) L310G mutant crystal structure | | 分子名称: | ACETATE ION, CALCIUM ION, Calmodulin, ... | | 著者 | Pylypenko, O, Song, L, Sweeney, L.H, Houdusse, A. | | 登録日 | 2010-01-05 | | 公開日 | 2010-12-08 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Role of insert i of myosin VI in modulating nucleotide affinity
To be Published
|
|
5HB1
 
 | |
3LBL
 
 | | Structure of human MDM2 protein in complex with Mi-63-analog | | 分子名称: | (2'R,3R,4'R,5'R)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-N-(2-morpholin-4-ylethyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide, E3 ubiquitin-protein ligase Mdm2 | | 著者 | Popowicz, G.M, Czarna, A, Wolf, S, Holak, T.A. | | 登録日 | 2010-01-08 | | 公開日 | 2010-03-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structures of low molecular weight inhibitors bound to MDMX and MDM2 reveal new approaches for p53-MDMX/MDM2 antagonist drug discovery
Cell Cycle, 9, 2010
|
|
5HB7
 
 | |
5HBE
 
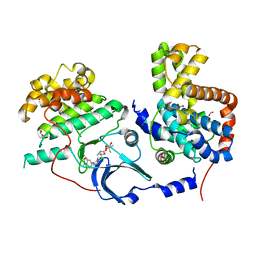 | | CDK8-CYCC IN COMPLEX WITH 8-[3-Chloro-5-(1-methyl-2,2-dioxo-2, 3-dihydro-1H-2l6-benzo[c]isothiazol-5-yl)-pyridin- 4-yl]-1-oxa-3,8-diaza-spiro[4.5]decan-2-one | | 分子名称: | 1,2-ETHANEDIOL, 8-[3-chloranyl-5-[1-methyl-2,2-bis(oxidanylidene)-3~{H}-2,1-benzothiazol-5-yl]pyridin-4-yl]-1-oxa-3,8-diazaspiro[4.5]decan-2-one, Cyclin-C, ... | | 著者 | Musil, D, Blagg, J, Mallinger, A. | | 登録日 | 2015-12-31 | | 公開日 | 2016-02-03 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Discovery of Potent, Selective, and Orally Bioavailable Small-Molecule Modulators of the Mediator Complex-Associated Kinases CDK8 and CDK19.
J.Med.Chem., 59, 2016
|
|
7NK4
 
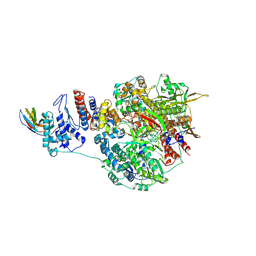 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8203 core | | 分子名称: | Nanobody 8203, Polymerase acidic protein, Polymerase basic protein 2,Polymerase basic protein 2, ... | | 著者 | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | 登録日 | 2021-02-17 | | 公開日 | 2021-12-01 | | 最終更新日 | 2022-02-09 | | 実験手法 | ELECTRON MICROSCOPY (5.32 Å) | | 主引用文献 | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
