6HUH
 
 | | CRYSTAL STRUCTURE OF OXA-427 class D BETA-LACTAMASE | | 分子名称: | Beta-lactamase, SULFATE ION | | 著者 | Zavala, A, Retailleau, P, Bogaerts, P, Glupczynski, Y, Naas, T, Iorga, B. | | 登録日 | 2018-10-08 | | 公開日 | 2019-10-30 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.78 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF CMY-OXA-427-HisTag BETA-LACTAMASE
To be published
|
|
4MO1
 
 | | Crystal structure of antitermination protein Q from bacteriophage lambda. Northeast Structural Genomics Consortium target OR18A. | | 分子名称: | Antitermination protein Q, BROMIDE ION, CHLORIDE ION, ... | | 著者 | Vorobiev, S, Su, M, Nickels, B, Seetharaman, J, Sahdev, S, Xiao, R, Kogan, S, Maglaqui, M, Wang, D, Everett, J.K, Acton, T.B, Ebright, R.H, Montelione, G.T, Hunt, J, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2013-09-11 | | 公開日 | 2013-09-25 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.099 Å) | | 主引用文献 | Crystal structure of antitermination protein Q from bacteriophage lambda.
To be Published
|
|
4MRW
 
 | | Crystal structure of PDE10A2 with fragment ZT0120 (7-chloroquinolin-4-ol) | | 分子名称: | 7-chloroquinolin-4-ol, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | 著者 | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, NIenaber, V. | | 登録日 | 2013-09-17 | | 公開日 | 2014-05-14 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
5A4V
 
 | | AtGSTF2 from Arabidopsis thaliana in complex with quercetin | | 分子名称: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, ACETATE ION, GLUTATHIONE S-TRANSFERASE F2 | | 著者 | Ahmad, L, Rylott, E, Bruce, N.C, Edwards, R, Grogan, G. | | 登録日 | 2015-06-15 | | 公開日 | 2016-06-29 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Structural evidence for Arabidopsis glutathione transferase AtGSTF2 functioning as a transporter of small organic ligands.
FEBS Open Bio, 7, 2017
|
|
5A7I
 
 | | Crystal structure of INPP5B in complex with biphenyl 3,3',4,4',5,5'- hexakisphosphate | | 分子名称: | Biphenyl 3,3',4,4',5,5'-hexakisphosphate, CHLORIDE ION, GLYCEROL, ... | | 著者 | Tresaugues, L, Mills, S.J, Silvander, C, Cozier, G, Potter, B.V.L, Norldund, P. | | 登録日 | 2015-07-06 | | 公開日 | 2016-04-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Crystal Structures of Type-II Inositol Polyphosphate 5-Phosphatase Inpp5B with Synthetic Inositol Polyphosphate Surrogates Reveal New Mechanistic Insights for the Inositol 5-Phosphatase Family.
Biochemistry, 55, 2016
|
|
5G4J
 
 | | Phospholyase A1RDF1 from Arthrobacter in complex with phosphoethanolamine | | 分子名称: | PUTATIVE AMINOTRANSFERASE CLASS III PROTEIN, SODIUM ION, {5-hydroxy-6-methyl-4-[(E)-{[2-(phosphonooxy)ethyl]imino}methyl]pyridin-3-yl}methyl dihydrogen phosphate | | 著者 | Cuetos, A, Tuan, A.N, Mangas Sanchez, J, Grogan, G. | | 登録日 | 2016-05-13 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Structural Basis for Phospholyase Activity of a Class III Transaminase Homologue.
Chembiochem, 17, 2016
|
|
4MSE
 
 | | Crystal structure of PDE10A2 with fragment ZT1597 (2-({[(2S)-2-methyl-2,3-dihydro-1,3-benzothiazol-5-yl]oxy}methyl)quinoline) | | 分子名称: | 2-{[(2-methyl-1,3-benzothiazol-5-yl)oxy]methyl}quinoline, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | 著者 | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | 登録日 | 2013-09-18 | | 公開日 | 2014-05-14 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
5GJI
 
 | | PI3K p85 N-terminal SH2 domain/CD28-derived peptide complex | | 分子名称: | GLYCEROL, Phosphatidylinositol 3-kinase regulatory subunit alpha, SULFATE ION, ... | | 著者 | Inaba, S, Numoto, N, Morii, H, Ogawa, S, Ikura, T, Abe, R, Ito, N, Oda, M. | | 登録日 | 2016-06-30 | | 公開日 | 2016-12-14 | | 最終更新日 | 2017-05-10 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Crystal Structures and Thermodynamic Analysis Reveal Distinct Mechanisms of CD28 Phosphopeptide Binding to the Src Homology 2 (SH2) Domains of Three Adaptor Proteins
J. Biol. Chem., 292, 2017
|
|
5G6S
 
 | | Imine reductase from Aspergillus oryzae in complex with NADP(H) and (R)-rasagiline | | 分子名称: | IMINE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, RASAGILINE | | 著者 | Man, H, Aleku, G, Turner, N.J, Grogan, G. | | 登録日 | 2016-06-23 | | 公開日 | 2017-05-31 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | A reductive aminase from Aspergillus oryzae.
Nat Chem, 9, 2017
|
|
5GIS
 
 | | Crystal structure of a Fab fragment with its ligand peptide | | 分子名称: | Heavy chain of Fab fragment, Light chain of Fab fragment, SULFATE ION, ... | | 著者 | Kitago, Y, Kaneko, K.K, Ogasawara, S, Kato, Y, Takagi, J. | | 登録日 | 2016-06-24 | | 公開日 | 2016-09-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Structural basis for multi-specific peptide recognition by the anti-IDH1/2 monoclonal antibody, MsMab-1.
Biochem. Biophys. Res. Commun., 478, 2016
|
|
5GJH
 
 | | Gads SH2 domain/CD28-derived peptide complex | | 分子名称: | GRB2-related adapter protein 2, T-cell-specific surface glycoprotein CD28 | | 著者 | Inaba, S, Numoto, N, Morii, H, Ogawa, S, Ikura, T, Abe, R, Ito, N, Oda, M. | | 登録日 | 2016-06-30 | | 公開日 | 2016-12-14 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Crystal Structures and Thermodynamic Analysis Reveal Distinct Mechanisms of CD28 Phosphopeptide Binding to the Src Homology 2 (SH2) Domains of Three Adaptor Proteins
J. Biol. Chem., 292, 2017
|
|
4XZY
 
 | | Crystal structure of dipeptidyl peptidase 11 (DPP11) from Porphyromonas gingivalis | | 分子名称: | GLYCEROL, Peptidase S46 | | 著者 | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | 登録日 | 2015-02-05 | | 公開日 | 2015-07-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural and mutational analyses of dipeptidyl peptidase 11 from Porphyromonas gingivalis reveal the molecular basis for strict substrate specificity.
Sci Rep, 5, 2015
|
|
5G5A
 
 | |
4Y01
 
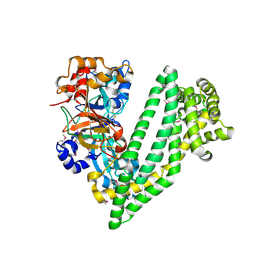 | | Crystal structure of dipeptidyl peptidase 11 (DPP11) from Porphyromonas gingivalis | | 分子名称: | GLYCEROL, Peptidase S46 | | 著者 | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | 登録日 | 2015-02-05 | | 公開日 | 2015-07-15 | | 最終更新日 | 2020-02-05 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | Structural and mutational analyses of dipeptidyl peptidase 11 from Porphyromonas gingivalis reveal the molecular basis for strict substrate specificity.
Sci Rep, 5, 2015
|
|
5C5G
 
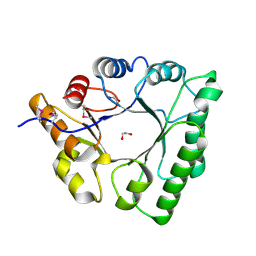 | |
5C5V
 
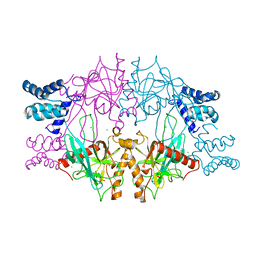 | | Recombinant Inorganic Pyrophosphatase from T brucei brucei | | 分子名称: | 1,2-ETHANEDIOL, Acidocalcisomal pyrophosphatase, BROMIDE ION, ... | | 著者 | Jamwal, A, Yogavel, M, Sharma, A. | | 登録日 | 2015-06-22 | | 公開日 | 2015-11-04 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural and Functional Highlights of Vacuolar Soluble Protein 1 from Pathogen Trypanosoma brucei brucei
J.Biol.Chem., 290, 2015
|
|
6I6S
 
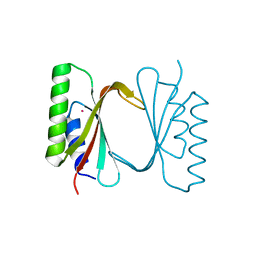 | | Circular permutant of ribosomal protein S6, adding 9aa to C terminal of P68-69, L75A mutant | | 分子名称: | 30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6, POTASSIUM ION, SODIUM ION | | 著者 | Wang, H, Logan, D.T, Oliveberg, M. | | 登録日 | 2018-11-15 | | 公開日 | 2019-11-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Exposing the distinctive modular behavior of beta-strands and alpha-helices in folded proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4Y02
 
 | | Crystal structure of dipeptidyl peptidase 11 (DPP11) from Porphyromonas gingivalis (Ground) | | 分子名称: | GLYCEROL, POTASSIUM ION, Peptidase S46 | | 著者 | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | 登録日 | 2015-02-05 | | 公開日 | 2015-07-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Structural and mutational analyses of dipeptidyl peptidase 11 from Porphyromonas gingivalis reveal the molecular basis for strict substrate specificity.
Sci Rep, 5, 2015
|
|
6IC7
 
 | | Human cathepsin-C in complex with dipeptidyl cyclopropyl nitrile inhibitor 3 | | 分子名称: | 1-azanyl-~{N}-[(1~{R},2~{R})-1-cyano-2-[4-[4-(4-methylpiperazin-1-yl)sulfonylphenyl]phenyl]cyclopropyl]cyclohexane-1-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | 著者 | Hakansson, M, Logan, D.T, Korkmaz, B, Lesner, A, Wysocka, M, Gieldon, A, Gauthier, F, Jenne, D, Lauritzen, C, Pedersen, J. | | 登録日 | 2018-12-02 | | 公開日 | 2019-04-24 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-based design and in vivo anti-arthritic activity evaluation of a potent dipeptidyl cyclopropyl nitrile inhibitor of cathepsin C.
Biochem. Pharmacol., 164, 2019
|
|
4LLP
 
 | | Crystal structure of PDE10A2 with fragment ZT401 | | 分子名称: | 4-amino-2-methylphenol, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | 著者 | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | 登録日 | 2013-07-09 | | 公開日 | 2014-02-26 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
4LM4
 
 | | Crystal structure of PDE10A2 with fragment ZT902 | | 分子名称: | NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A, quinazolin-4(1H)-one | | 著者 | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | 登録日 | 2013-07-10 | | 公開日 | 2014-02-26 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
6HGF
 
 | | Crystal structure of Alpha1-antichymotrypsin variant NewBG-II: a new binding globulin in complex with cortisol | | 分子名称: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, 1,2-ETHANEDIOL, Alpha-1-antichymotrypsin | | 著者 | Schmidt, K, Muller, Y.A. | | 登録日 | 2018-08-23 | | 公開日 | 2019-05-29 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.651 Å) | | 主引用文献 | NewBG: A surrogate corticosteroid-binding globulin with an unprecedentedly high ligand release efficacy.
J.Struct.Biol., 207, 2019
|
|
6HGH
 
 | |
6I07
 
 | | Crystal structure of EpCAM in complex with scFv | | 分子名称: | Epithelial cell adhesion molecule, GLYCEROL, Single chain Fv | | 著者 | Casaletto, J.B, Geddie, M.L, Abu-Yousif, A.O, Masson, K, Fulgham, A, Boudot, A, Maiwald, T, Kearns, J.D, Kohli, N, Su, S, Razlog, M, Raue, A, Kalra, A, Hakansson, M, Logan, D.T, Welin, M, Chattopadhyay, S, Harms, B.D, Nielsen, U.B, Schoeberl, B, Lugovskoy, A.A, MacBeath, G. | | 登録日 | 2018-10-25 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | MM-131, a bispecific anti-Met/EpCAM mAb, inhibits HGF-dependent and HGF-independent Met signaling through concurrent binding to EpCAM.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4LNY
 
 | | Crystal Structure of Engineered Protein, Northeast Structural Genomics Consortium Target OR422 | | 分子名称: | CADMIUM ION, CHLORIDE ION, Engineered Protein OR422 | | 著者 | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Sahdev, S, Xiao, R, Maglaqui, M, Kogan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2013-07-12 | | 公開日 | 2013-08-07 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.929 Å) | | 主引用文献 | Crystal Structure of Engineered Protein OR422.
To be Published
|
|
