5FGB
 
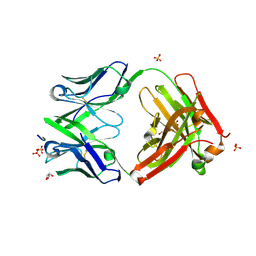 | | Three dimensional structure of broadly neutralizing human anti - Hepatitis C virus (HCV) glycoprotein E2 Fab fragment HC33.4 | | 分子名称: | Anti-HCV E2 Fab HC84-1 heavy chain, Anti-HCV E2 Fab HC84-1 light chain, GLYCEROL, ... | | 著者 | Girard-Blanc, C, Rey, F.A, Krey, T. | | 登録日 | 2015-12-20 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Antibody Response to Hypervariable Region 1 Interferes with Broadly Neutralizing Antibodies to Hepatitis C Virus.
J.Virol., 90, 2016
|
|
6ENR
 
 | |
5JPH
 
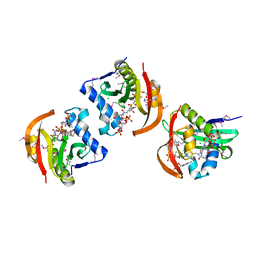 | | Structure of a GNAT acetyltransferase SACOL1063 from Staphylococcus aureus in complex with CoA | | 分子名称: | Acetyltransferase SACOL1063, CHLORIDE ION, COENZYME A | | 著者 | Majorek, K.A, Osinski, T, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-05-03 | | 公開日 | 2016-06-29 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Insight into the 3D structure and substrate specificity of previously uncharacterized GNAT superfamily acetyltransferases from pathogenic bacteria.
Biochim.Biophys.Acta, 1865, 2016
|
|
5JQ4
 
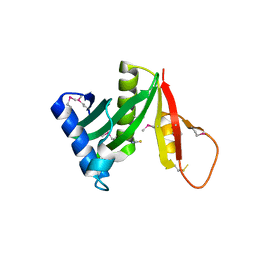 | | Structure of a GNAT acetyltransferase SACOL1063 from Staphylococcus aureus | | 分子名称: | 1,2-ETHANEDIOL, Acetyltransferase SACOL1063, CHLORIDE ION, ... | | 著者 | Majorek, K.A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-05-04 | | 公開日 | 2016-06-29 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Insight into the 3D structure and substrate specificity of previously uncharacterized GNAT superfamily acetyltransferases from pathogenic bacteria.
Biochim.Biophys.Acta, 1865, 2016
|
|
4AGD
 
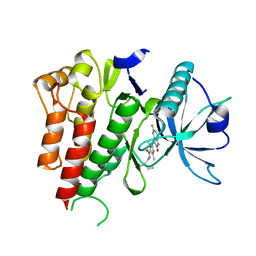 | | CRYSTAL STRUCTURE OF VEGFR2 (JUXTAMEMBRANE AND KINASE DOMAINS) IN COMPLEX WITH SUNITINIB (SU11248) (N-2-diethylaminoethyl)-5-((Z)-(5- fluoro-2-oxo-1H-indol-3-ylidene)methyl)-2,4-dimethyl-1H-pyrrole-3- carboxamide) | | 分子名称: | N-[2-(diethylamino)ethyl]-5-[(Z)-(5-fluoro-2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-2,4-dimethyl-1H-pyrrole-3-carbo xamide, VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR 2 | | 著者 | McTigue, M, Deng, Y, Ryan, K, Brooun, A, Diehl, W, Stewart, A. | | 登録日 | 2012-01-26 | | 公開日 | 2012-09-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Molecular Conformations, Interactions, and Properties Associated with Drug Efficiency and Clinical Performance Among Vegfr Tk Inhibitors.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6GSD
 
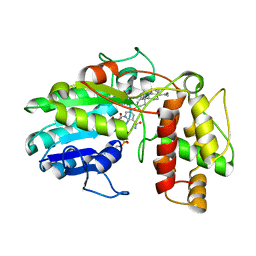 | | Plantago Major multifunctional oxidoreductase in complex with progesterone and NADP+ | | 分子名称: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROGESTERONE, Progesterone 5-beta-reductase | | 著者 | Fellows, R, Silva, C, Russo, C.M, Lee, S.G, Jez, J.M, Chisholm, J.D, Zubieta, C, Nanao, M. | | 登録日 | 2018-06-14 | | 公開日 | 2018-10-17 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | A multisubstrate reductase from Plantago major: structure-function in the short chain reductase superfamily.
Sci Rep, 8, 2018
|
|
3KW0
 
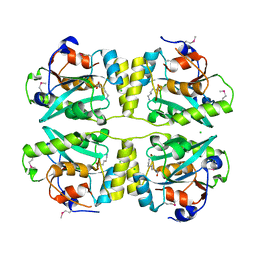 | |
2VNG
 
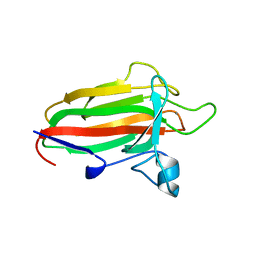 | | Family 51 carbohydrate binding module from a family 98 glycoside hydrolase produced by Clostridium perfringens in complex with blood group A-trisaccharide ligand. | | 分子名称: | CALCIUM ION, CPE0329, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose | | 著者 | Gregg, K.J, Finn, R, Abbott, D.W, Boraston, A.B. | | 登録日 | 2008-02-04 | | 公開日 | 2008-02-19 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Divergent Modes of Glycan Recognition by a New Family of Carbohydrate-Binding Modules
J.Biol.Chem., 283, 2008
|
|
4AG8
 
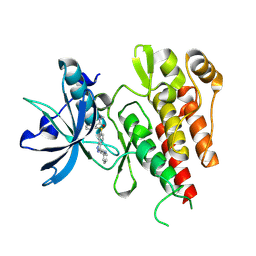 | | CRYSTAL STRUCTURE OF THE VEGFR2 KINASE DOMAIN IN COMPLEX WITH AXITINIB (AG-013736) (N-Methyl-2-(3-((E)-2-pyridin-2-yl-vinyl)-1H- indazol-6-ylsulfanyl)-benzamide) | | 分子名称: | AXITINIB, VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR 2 | | 著者 | McTigue, M, Wickersham, J, Pinko, C, Kania, R.S, Bender, S. | | 登録日 | 2012-01-24 | | 公開日 | 2012-09-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Molecular Conformations, Interactions, and Properties Associated with Drug Efficiency and Clinical Performance Among Vegfr Tk Inhibitors.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AGC
 
 | | CRYSTAL STRUCTURE OF VEGFR2 (JUXTAMEMBRANE AND KINASE DOMAINS) IN COMPLEX WITH AXITINIB (AG-013736) (N-Methyl-2-(3-((E)-2-pyridin-2-yl- vinyl)-1H-indazol-6-ylsulfanyl)-benzamide) | | 分子名称: | AXITINIB, VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR 2 | | 著者 | Mctigue, M, Deng, Y, Ryan, K, Brooun, A, Diehl, W, Kania, R.S. | | 登録日 | 2012-01-26 | | 公開日 | 2012-09-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Molecular Conformations, Interactions, and Properties Associated with Drug Efficiency and Clinical Performance Among Vegfr Tk Inhibitors.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4ASD
 
 | | Crystal Structure of VEGFR2 (Juxtamembrane and Kinase Domains) in Complex with SORAFENIB (BAY 43-9006) | | 分子名称: | 4-{4-[({[4-CHLORO-3-(TRIFLUOROMETHYL)PHENYL]AMINO}CARBONYL)AMINO]PHENOXY}-N-METHYLPYRIDINE-2-CARBOXAMIDE, VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR 2 | | 著者 | McTigue, M, Deng, Y, Ryan, K, Brooun, A, Diehl, W, Stewart, A. | | 登録日 | 2012-04-30 | | 公開日 | 2012-09-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Molecular Conformations, Interactions, and Properties Associated with Drug Efficiency and Clinical Performance Among Vegfr Tk Inhibitors.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VNE
 
 | |
6EEJ
 
 | | Streptomyces bingchenggensis Aldolase-Dehydratase in covalent complex with dienone product. | | 分子名称: | (3E,5E)-6-(4-nitrophenyl)-2-oxohexa-3,5-dienoic acid, Acetoacetate decarboxylase, FORMIC ACID, ... | | 著者 | Mydy, L.S, Silvaggi, N.R. | | 登録日 | 2018-08-14 | | 公開日 | 2019-08-14 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.892 Å) | | 主引用文献 | Mechanistic Studies of theStreptomyces bingchenggensisAldolase-Dehydratase: Implications for Substrate and Reaction Specificity in the Acetoacetate Decarboxylase-like Superfamily.
Biochemistry, 58, 2019
|
|
5EL1
 
 | |
5EMU
 
 | |
6EP9
 
 | |
1AUP
 
 | | GLUTAMATE DEHYDROGENASE | | 分子名称: | NAD-SPECIFIC GLUTAMATE DEHYDROGENASE | | 著者 | Baker, P.J, Waugh, M.L, Stillman, T.J, Turnbull, A.P, Rice, D.W. | | 登録日 | 1997-09-01 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Determinants of substrate specificity in the superfamily of amino acid dehydrogenases.
Biochemistry, 36, 1997
|
|
5NRA
 
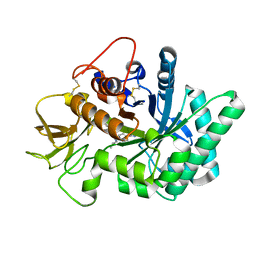 | | Crystal structure of human chitotriosidase-1 (hCHIT) catalytic domain in complex with compound 7g | | 分子名称: | 1-(5-azanyl-4~{H}-1,2,4-triazol-3-yl)-~{N}-[2-(4-bromophenyl)ethyl]-~{N}-(2-methylpropyl)piperidin-4-amine, Chitotriosidase-1, GLYCEROL | | 著者 | Mazur, M, Olczak, J, Olejniczak, S, Koralewski, R, Czestkowski, W, Jedrzejczak, A, Golab, J, Dzwonek, K, Dymek, B, Sklepkiewicz, P, Zagozdzon, A, Noonan, T, Mahboubi, K, Conway, B, Sheeler, R, Beckett, P, Hungerford, W.M, Podjarny, A, Mitschler, A, Cousido-Siah, A, Fadel, F, Golebiowski, A. | | 登録日 | 2017-04-22 | | 公開日 | 2018-03-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.267 Å) | | 主引用文献 | Targeting Acidic Mammalian chitinase Is Effective in Animal Model of Asthma.
J. Med. Chem., 61, 2018
|
|
6IFY
 
 | | Type III-A Csm complex, Cryo-EM structure of Csm-CTR1 | | 分子名称: | CTR1, Type III-A CRISPR-associated RAMP protein Csm3, Type III-A CRISPR-associated RAMP protein Csm4, ... | | 著者 | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | 登録日 | 2018-09-21 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
6IFL
 
 | | Cryo-EM structure of type III-A Csm-NTR complex | | 分子名称: | NTR, Type III-A CRISPR-associated RAMP protein Csm3, Type III-A CRISPR-associated RAMP protein Csm4, ... | | 著者 | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | 登録日 | 2018-09-20 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.16 Å) | | 主引用文献 | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
1JPM
 
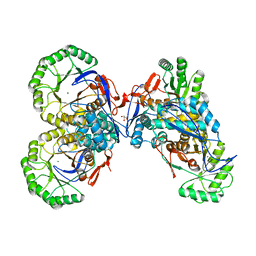 | | L-Ala-D/L-Glu Epimerase | | 分子名称: | GLYCEROL, L-Ala-D/L-Glu Epimerase, MAGNESIUM ION | | 著者 | Gulick, A.M, Schmidt, D.M.Z, Gerlt, J.A, Rayment, I. | | 登録日 | 2001-08-02 | | 公開日 | 2001-12-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Evolution of enzymatic activities in the enolase superfamily: crystal structures of the L-Ala-D/L-Glu epimerases from Escherichia coli and Bacillus subtilis.
Biochemistry, 40, 2001
|
|
3VNF
 
 | |
5NLV
 
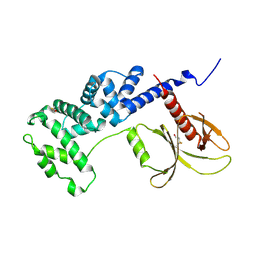 | | Brag2 Sec7-PH (390-763) | | 分子名称: | IQ motif and SEC7 domain-containing protein 1 | | 著者 | Nawrotek, A, Cherfils, J. | | 登録日 | 2017-04-05 | | 公開日 | 2017-09-27 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Multiple interactions between an Arf/GEF complex and charged lipids determine activation kinetics on the membrane.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5MG8
 
 | | Crystal structure of the S.pombe Smc5/6 hinge domain | | 分子名称: | GLYCEROL, SULFATE ION, Structural maintenance of chromosomes protein 5, ... | | 著者 | Alt, A, Pearl, L.H, Oliver, A.W. | | 登録日 | 2016-11-21 | | 公開日 | 2017-02-08 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Specialized interfaces of Smc5/6 control hinge stability and DNA association.
Nat Commun, 8, 2017
|
|
5MAL
 
 | |
