3QI7
 
 | |
4FUU
 
 | |
4G0S
 
 | |
3LOU
 
 | |
4FVR
 
 | |
3LEW
 
 | |
2R3E
 
 | |
8EAQ
 
 | | Structure of the full-length IP3R1 channel determined at high Ca2+ | | 分子名称: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, CALCIUM ION, Inositol 1,4,5-trisphosphate receptor type 1, ... | | 著者 | Fan, G, Baker, M.R, Terry, L.E, Arige, V, Chen, M, Seryshev, A.B, Baker, M.L, Ludtke, S.J, Yule, D.I, Serysheva, I.I. | | 登録日 | 2022-08-29 | | 公開日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (3.26 Å) | | 主引用文献 | Conformational motions and ligand-binding underlying gating and regulation in IP 3 R channel.
Nat Commun, 13, 2022
|
|
3LWC
 
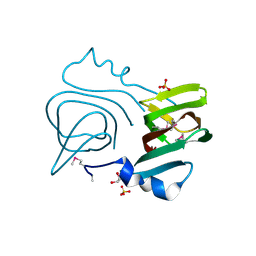 | |
8EAR
 
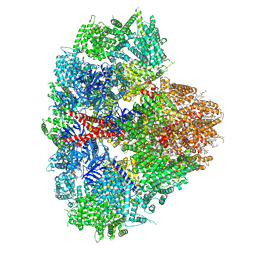 | | Structure of the full-length IP3R1 channel determined in the presence of Calcium/IP3/ATP | | 分子名称: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | 著者 | Fan, G, Baker, M.R, Terry, L.E, Arige, V, Chen, M, Seryshev, A.B, Baker, M.L, Ludtke, S.J, Yule, D.I, Serysheva, I.I. | | 登録日 | 2022-08-29 | | 公開日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Conformational motions and ligand-binding underlying gating and regulation in IP 3 R channel.
Nat Commun, 13, 2022
|
|
4GBM
 
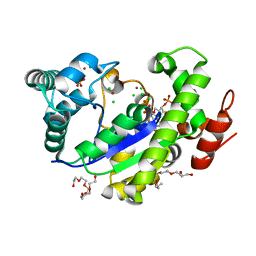 | |
4GB9
 
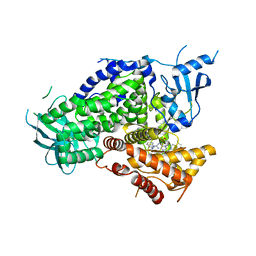 | | Potent and Highly Selective Benzimidazole Inhibitors of PI3K-delta | | 分子名称: | 2-[1-({2-[2-(dimethylamino)-1H-benzimidazol-1-yl]-9-methyl-6-(morpholin-4-yl)-9H-purin-8-yl}methyl)piperidin-4-yl]propan-2-ol, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | 著者 | Murray, J.M. | | 登録日 | 2012-07-26 | | 公開日 | 2012-08-22 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.438 Å) | | 主引用文献 | Potent and highly selective benzimidazole inhibitors of PI3-kinase delta.
J.Med.Chem., 55, 2012
|
|
3LJY
 
 | |
2QWM
 
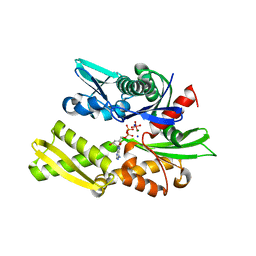 | | Crystal structure of bovine hsc70 (1-394aa)in the ADP*Vi state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, Heat shock cognate 71 kDa protein, ... | | 著者 | Jiang, J, Maes, E.G, Wang, L, Taylor, A.B, Hinck, A.P, Lafer, E.M, Sousa, R. | | 登録日 | 2007-08-10 | | 公開日 | 2007-12-18 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Structural basis of J cochaperone binding and regulation of Hsp70.
Mol.Cell, 28, 2007
|
|
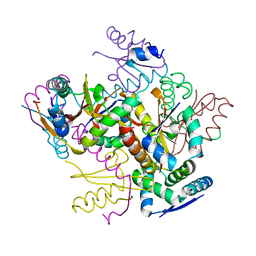
4FXE
 
 | |
3LMZ
 
 | |
2R4T
 
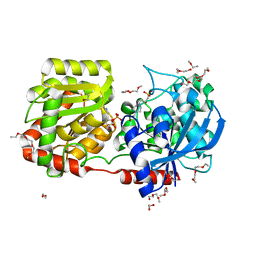 | | Crystal Structure of Wild-type E.coli GS in Complex with ADP and Glucose(wtGSc) | | 分子名称: | (2R)-2-hydroxy-3-[4-(2-hydroxyethyl)piperazin-1-yl]propane-1-sulfonic acid, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Sheng, F, Geiger, J. | | 登録日 | 2007-09-01 | | 公開日 | 2008-09-09 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.258 Å) | | 主引用文献 | The crystal structures of the open and catalytically competent closed conformation of Escherichia coli glycogen synthase.
J.Biol.Chem., 284, 2009
|
|
2R7O
 
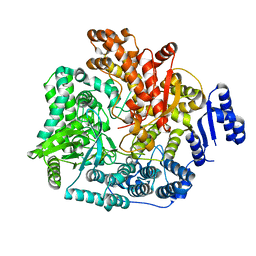 | | Crystal Structure of VP1 apoenzyme of Rotavirus SA11 (N-terminal hexahistidine-tagged) | | 分子名称: | RNA-dependent RNA polymerase | | 著者 | Lu, X, Harrison, S.C, Tao, Y.J, Patton, J.T, Nibert, M.L. | | 登録日 | 2007-09-09 | | 公開日 | 2008-07-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1.
Structure, 16, 2008
|
|
2R7S
 
 | | Crystal Structure of Rotavirus SA11 VP1 / RNA (UGUGCC) complex | | 分子名称: | PHOSPHATE ION, RNA (5'-R(*UP*GP*UP*GP*CP*C)-3'), RNA-dependent RNA polymerase | | 著者 | Lu, X, Harrison, S.C, Tao, Y.J, Patton, J.T, Nibert, M.L. | | 登録日 | 2007-09-10 | | 公開日 | 2008-07-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.24 Å) | | 主引用文献 | Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1.
Structure, 16, 2008
|
|
4GBK
 
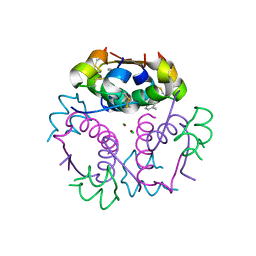 | | Crystal structure of aspart insulin at pH 8.5 | | 分子名称: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | 著者 | Lima, L.M.T.R, Favero-Retto, M.P, Palmieri, L.C. | | 登録日 | 2012-07-27 | | 公開日 | 2013-06-12 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | A T3R3 hexamer of the human insulin variant B28Asp.
Biophys.Chem., 173, 2013
|
|
2RC4
 
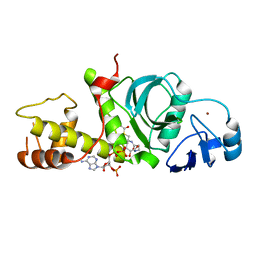 | | Crystal Structure of the HAT domain of the human MOZ protein | | 分子名称: | ACETYL COENZYME *A, Histone acetyltransferase MYST3, ZINC ION | | 著者 | Holbert, M.A, Sikorski, T, Snowflack, D, Marmorstein, R. | | 登録日 | 2007-09-19 | | 公開日 | 2007-11-13 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The human monocytic leukemia zinc finger histone acetyltransferase domain contains DNA-binding activity implicated in chromatin targeting.
J.Biol.Chem., 282, 2007
|
|
3LLC
 
 | |
3LM2
 
 | |
3LN3
 
 | |
2R7V
 
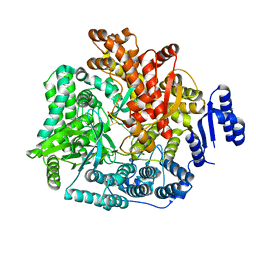 | | Crystal Structure of Rotavirus SA11 VP1/RNA (GGCUUU) Complex | | 分子名称: | RNA (5'-R(*G*GP*CP*UP*UP*U)-3'), RNA-dependent RNA polymerase | | 著者 | Lu, X, Harrison, S.C, Tao, Y.J, Patton, J.T, Nibert, M.L. | | 登録日 | 2007-09-10 | | 公開日 | 2008-07-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1.
Structure, 16, 2008
|
|
