7PJV
 
 | | Structure of the 70S-EF-G-GDP-Pi ribosome complex with tRNAs in hybrid state 1 (H1-EF-G-GDP-Pi) | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Petrychenko, V, Peng, B.Z, Schwarzer, A.C, Peske, F, Rodnina, M.V, Fischer, N. | | 登録日 | 2021-08-24 | | 公開日 | 2021-10-20 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural mechanism of GTPase-powered ribosome-tRNA movement.
Nat Commun, 12, 2021
|
|
7QG8
 
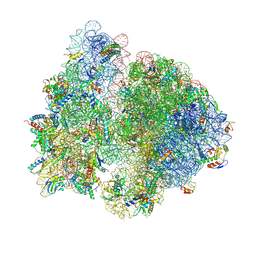 | | Structure of the collided E. coli disome - VemP-stalled 70S ribosome | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Kratzat, H, Buschauer, R, Berninghausen, O, Beckmann, R. | | 登録日 | 2021-12-07 | | 公開日 | 2022-03-16 | | 最終更新日 | 2022-03-30 | | 実験手法 | ELECTRON MICROSCOPY (3.97 Å) | | 主引用文献 | Ribosome collisions induce mRNA cleavage and ribosome rescue in bacteria.
Nature, 603, 2022
|
|
7QGR
 
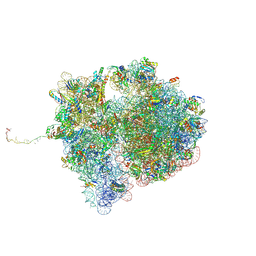 | | Structure of the SmrB-bound E. coli disome - collided 70S ribosome | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Kratzat, H, Buschauer, R, Berninghausen, O, Beckmann, R. | | 登録日 | 2021-12-09 | | 公開日 | 2022-06-22 | | 実験手法 | ELECTRON MICROSCOPY (5.7 Å) | | 主引用文献 | Ribosome collisions induce mRNA cleavage and ribosome rescue in bacteria.
Nature, 603, 2022
|
|
1MJC
 
 | |
1MM2
 
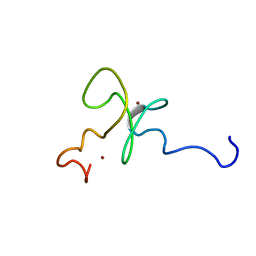 | | Solution structure of the 2nd PHD domain from Mi2b | | 分子名称: | Mi2-beta, ZINC ION | | 著者 | Kwan, A.H.Y, Gell, D.A, Verger, A, Crossley, M, Matthews, J.M, Mackay, J.P. | | 登録日 | 2002-09-02 | | 公開日 | 2003-07-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Engineering a Protein Scaffold from a PHD Finger
structure, 11, 2003
|
|
3L1Q
 
 | | The crystal structure of the undecamer d(TGGCCTTAAGG) | | 分子名称: | 5'-D(*TP*GP*GP*CP*CP*TP*TP*AP*AP*GP*G)-3' | | 著者 | Van Hecke, K. | | 登録日 | 2009-12-14 | | 公開日 | 2010-10-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Designing Triple Helical Fragments: The Crystal Structure of the Undecamer d(TGGCCTTAAGG) Mimicking T·AT Base Triplets
Cryst.Growth Des., 10, 2010
|
|
4UV9
 
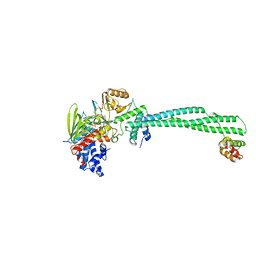 | | LSD1(KDM1A)-CoREST in complex with 1-Ethyl-Tranylcypromine | | 分子名称: | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S)-2,3,4-trihydroxy-5-[(4aS,10aS)-4a-[(1S,3E)-3-imino-1-phenylpentyl]-7,8-dimethyl-2,4-dioxo-1,3,4,4a,5,10a-hexahydrobenzo[g]pteridin-10(2H)-yl]pentyl dihydrogen diphosphate | | 著者 | Vianello, P, Botrugno, O, Cappa, A, Ciossani, G, Dessanti, P, Mai, A, Mattevi, A, Meroni, G, Minucci, S, Thaler, F, Tortorici, M, Trifiro, P, Valente, S, Villa, M, Varasi, M, Mercurio, C. | | 登録日 | 2014-08-05 | | 公開日 | 2014-09-10 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Synthesis, Biological Activity and Mechanistic Insights of 1-Substituted Cyclopropylamine Derivatives: A Novel Class of Irreversible Inhibitors of Histone Demethylase Kdm1A.
Eur.J.Med.Chem., 86C, 2014
|
|
2QNP
 
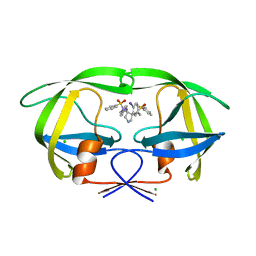 | | HIV-1 Protease in complex with a iodo decorated pyrrolidine-based inhibitor | | 分子名称: | CHLORIDE ION, Gag-Pol polyprotein (Pr160Gag-Pol), N,N'-(3S,4S)-pyrrolidine-3,4-diylbis[N-(4-iodobenzyl)benzenesulfonamide] | | 著者 | Boettcher, J, Blum, A, Heine, A, Diederich, W.E, Klebe, G. | | 登録日 | 2007-07-19 | | 公開日 | 2008-04-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | Structure-Guided Design of C2-Symmetric HIV-1 Protease Inhibitors Based on a Pyrrolidine Scaffold.
J.Med.Chem., 51, 2008
|
|
2QNQ
 
 | | HIV-1 Protease in complex with a chloro decorated pyrrolidine-based inhibitor | | 分子名称: | CHLORIDE ION, Gag-Pol polyprotein (Pr160Gag-Pol), N,N'-(3S,4S)-pyrrolidine-3,4-diylbis(N-benzyl-2-chlorobenzenesulfonamide) | | 著者 | Boettcher, J, Blum, A, Heine, A, Diederich, W.E, Klebe, G. | | 登録日 | 2007-07-19 | | 公開日 | 2008-04-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure-Guided Design of C2-Symmetric HIV-1 Protease Inhibitors Based on a Pyrrolidine Scaffold.
J.Med.Chem., 51, 2008
|
|
6ULG
 
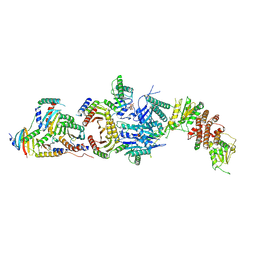 | | Cryo-EM structure of the FLCN-FNIP2-Rag-Ragulator complex | | 分子名称: | Folliculin, Folliculin-interacting protein 2, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Shen, K, Rogala, K.B, Yu, Z.H, Sabatini, D.M. | | 登録日 | 2019-10-08 | | 公開日 | 2019-11-20 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (3.31 Å) | | 主引用文献 | Cryo-EM Structure of the Human FLCN-FNIP2-Rag-Ragulator Complex.
Cell, 179, 2019
|
|
6UX2
 
 | |
9EMA
 
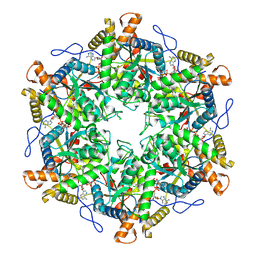 | | RUVBL1/2 in complex with ATP and CB-6644 inhibitor | | 分子名称: | 5-chloranyl-2-ethoxy-4-fluoranyl-~{N}-[4-[[3-(methoxymethyl)-1-oxidanylidene-6,7-dihydro-5~{H}-pyrazolo[1,2-a][1,2]benzodiazepin-2-yl]amino]-2,2-dimethyl-4-oxidanylidene-butyl]benzamide, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Lopez-Perrote, A, Llorca, O, Garcia-Martin, C. | | 登録日 | 2024-03-07 | | 公開日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | Mechanism of allosteric inhibition of RUVBL1-RUVBL2 by the small-molecule CB-6644
Cell Rep Phys Sci, 2024
|
|
3BRO
 
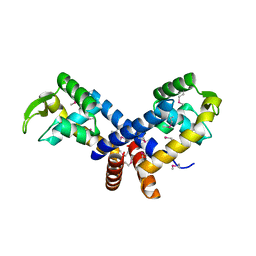 | | Crystal structure of the transcription regulator MarR from Oenococcus oeni PSU-1 | | 分子名称: | CHLORIDE ION, GLYCEROL, Transcriptional regulator | | 著者 | Kim, Y, Volkart, L, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2007-12-21 | | 公開日 | 2008-01-15 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Crystal Structure of the Transcription Regulator MarR from Oenococcus oeni PSU-1.
To be Published
|
|
6OF0
 
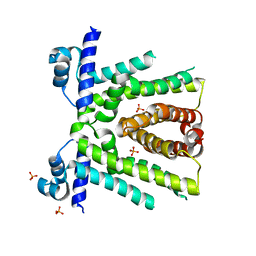 | | Structural basis for multidrug recognition and antimicrobial resistance by MTRR, an efflux pump regulator from Neisseria Gonorrhoeae | | 分子名称: | HTH-type transcriptional regulator MtrR, PHOSPHATE ION | | 著者 | Beggs, G.A, Kumaraswami, M, Shafer, W, Brennan, R.G. | | 登録日 | 2019-03-28 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural, Biochemical, and In Vivo Characterization of MtrR-Mediated Resistance to Innate Antimicrobials by the Human Pathogen Neisseria gonorrhoeae .
J.Bacteriol., 201, 2019
|
|
3CC2
 
 | |
3CCL
 
 | |
3CCV
 
 | |
3CCU
 
 | |
3CC7
 
 | |
3CCE
 
 | |
3CDH
 
 | | Crystal structure of the MarR family transcriptional regulator SPO1453 from Silicibacter pomeroyi DSS-3 | | 分子名称: | GLYCEROL, SULFATE ION, Transcriptional regulator, ... | | 著者 | Kim, Y, Volkart, L, Keigher, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-02-26 | | 公開日 | 2008-03-18 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Crystal structure of the MarR family transcriptional regulator SPO1453 from Silicibacter pomeroyi.
To be Published
|
|
3CC4
 
 | |
3CCM
 
 | |
3CCJ
 
 | |
3CCR
 
 | |
