9F43
 
 | |
9BZN
 
 | | High resolution structure of class A Beta-lactamase from Bordetella bronchiseptica RB50 | | 分子名称: | FORMIC ACID, SULFATE ION, class A Beta-lactamase, ... | | 著者 | Maltseva, N, Kim, Y, Endres, M, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2024-05-24 | | 公開日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | High resolution structure of class A Beta-lactamase from Bordetella bronchiseptica RB50
To Be Published
|
|
9C4B
 
 | | Second BAF53a of the human TIP60 complex | | 分子名称: | Actin-like protein 6A | | 著者 | Yang, Z, Mameri, A, Florez Ariza, A.J, Cote, J, Nogales, E. | | 登録日 | 2024-06-03 | | 公開日 | 2024-08-14 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural insights into the human NuA4/TIP60 acetyltransferase and chromatin remodeling complex.
Science, 2024
|
|
8W68
 
 | | Crystal structure of Q9PR55 at pH 6.0 (use NMR model) | | 分子名称: | Uncharacterized protein UU089.1 | | 著者 | Hsu, M.F, Ko, T.P, Huang, K.F, Chen, Y.R, Huang, J.S, Hsu, S.T.D. | | 登録日 | 2023-08-28 | | 公開日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure, dynamics, and stability of the smallest and most complex 7 1 protein knot.
J.Biol.Chem., 300, 2023
|
|
8WBC
 
 | |
9BXI
 
 | |
8XD4
 
 | |
8X5I
 
 | |
9EUB
 
 | | The FK1 domain of FKBP51 in complex with SAFit-analog 24e | | 分子名称: | Peptidyl-prolyl cis-trans isomerase FKBP5, [1-(2-hydroxyethyl)pyrazol-4-yl]methyl (2S)-1-[(2S)-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidine-2-carboxylate | | 著者 | Meyners, C, Buffa, V, Hausch, F. | | 登録日 | 2024-03-27 | | 公開日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | 1,4-Pyrazolyl-containing SAFit-analogues are selective FKBP51 inhibitors with improved ligand efficiency and drug-like profile.
Chemmedchem, 2024
|
|
9ATT
 
 | | Crystal structure of MERS 3CL protease in complex with a methylcyclohexyl 2-pyrrolidone inhibitor (R-enantiomer) | | 分子名称: | (1R,2S)-2-{[N-({[(2R)-1-(cyclohexylmethyl)-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[(2R)-1-(cyclohexylmethyl)-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | 著者 | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | 登録日 | 2024-02-27 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
8YBG
 
 | |
8VCE
 
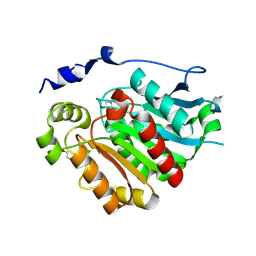 | | Crystal Structure of plant Carboxylesterase 20 | | 分子名称: | 1,2-ETHANEDIOL, IMIDAZOLE, Probable carboxylesterase 120 | | 著者 | Palayam, M, Shabek, N. | | 登録日 | 2023-12-14 | | 公開日 | 2024-08-07 | | 最終更新日 | 2024-08-14 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural insights into strigolactone catabolism by carboxylesterases reveal a conserved conformational regulation.
Nat Commun, 15, 2024
|
|
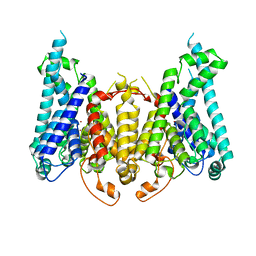
8W9V
 
 | |
9CIP
 
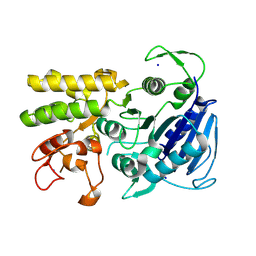 | | MicroED structure of the C11 cysteine protease clostripain | | 分子名称: | Clostripain, SODIUM ION | | 著者 | Ruma, Y.N, Bu, G, Hattne, J, Gonen, T. | | 登録日 | 2024-07-03 | | 公開日 | 2024-08-14 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (2.5 Å) | | 主引用文献 | MicroED structure of the C11 cysteine protease clostripain.
J Struct Biol X, 10, 2024
|
|
9EUS
 
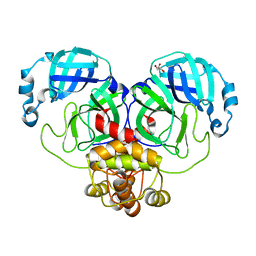 | | Mpro from SARS-CoV-2 with R298A mutation | | 分子名称: | GLYCEROL, Replicase polyprotein 1a | | 著者 | Plewka, J, Lis, K, Czarna, A, Pyrc, K, Kantyka, T, Chykunova, Y. | | 登録日 | 2024-03-28 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | SARS-CoV-2 M pro oligomerization as a potential target for therapy.
Int.J.Biol.Macromol., 267, 2024
|
|
9FTW
 
 | |
8VIJ
 
 | | Crystal structure of Shewanella benthica Group 1 truncated hemoglobin Y34F C51S C71S variant (cyanomet) | | 分子名称: | CYANIDE ION, Group 1 truncated hemoglobin, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Lecomte, J.T.J, Schlessman, J.L, Martinez, J.E, Schultz, T.D, Siegler, M.A, DelCampo, M, Le Magueres, P. | | 登録日 | 2024-01-04 | | 公開日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Crystal structure of Shewanella benthica Group 1 truncated hemoglobin Y34F C51S C71S variant in the cyanomet state
To be published
|
|
8XCR
 
 | |
9C8S
 
 | |
8WTB
 
 | |
8X7Z
 
 | |
9BKK
 
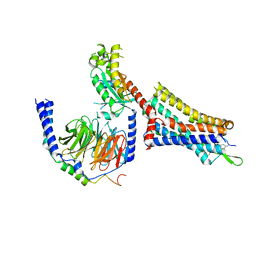 | | Cholecystokinin 1 receptor (CCK1R) sterol 7M mutant, Gq chimera (mGsqi) complex | | 分子名称: | Cholecystokinin receptor type A, Cholecystokinin-8, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Harikumar, K.G, Zhao, P, Cary, B.P, Xu, X, Desai, A.J, Mobbs, J.I, Toufaily, C, Furness, S.G.B, Christopoulos, A, Belousoff, M.J, Wootten, D, Sexton, P.M, Miller, L.J. | | 登録日 | 2024-04-29 | | 公開日 | 2024-05-22 | | 最終更新日 | 2024-08-14 | | 実験手法 | ELECTRON MICROSCOPY (2.51 Å) | | 主引用文献 | Cholesterol-dependent dynamic changes in the conformation of the type 1 cholecystokinin receptor affect ligand binding and G protein coupling.
Plos Biol., 22, 2024
|
|
8VX9
 
 | |
8W6V
 
 | |
9EM3
 
 | | OPR3 wild type in its monomeric form | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 12-oxophytodienoate reductase 3, FLAVIN MONONUCLEOTIDE | | 著者 | Bijelic, A, Macheroux, P, Kerschbaumer, B. | | 登録日 | 2024-03-07 | | 公開日 | 2024-08-14 | | 最終更新日 | 2024-08-21 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Analysis of homodimer formation in 12-oxophytodienoate reductase 3 in solutio and crystallo challenges the physiological role of the dimer.
Sci Rep, 14, 2024
|
|
