7NLC
 
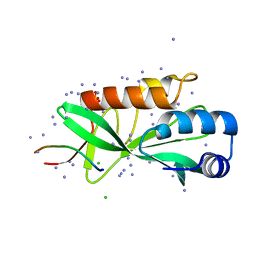 | | Crystallographic structure of human Tsg101 UEV domain in complex with a HEV ORF3 peptide | | 分子名称: | AMMONIUM ION, CHLORIDE ION, Protein ORF3, ... | | 著者 | Moschidi, D, Dupre, E, Villeret, V, Hanoulle, X. | | 登録日 | 2021-02-22 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.398 Å) | | 主引用文献 | Crystallographic structure of human Tsg101 UEV domain in complex with a HEV ORF3 peptide
To Be Published
|
|
7ZRA
 
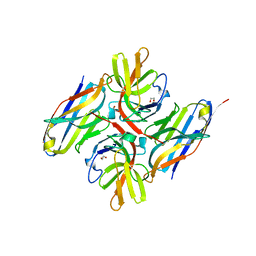 | | Crystal structure of E.coli LexA in complex with nanobody NbSOS1(Nb14497) | | 分子名称: | 1,2-ETHANEDIOL, LexA repressor, Nanobody NbSOS1 (Nb14497) | | 著者 | Maso, L, Vascon, F, Chinellato, M, Pardon, E, Steyaert, J, Angelini, A, Tondi, D, Cendron, L. | | 登録日 | 2022-05-04 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Nanobodies targeting LexA autocleavage disclose a novel suppression strategy of SOS-response pathway.
Structure, 30, 2022
|
|
6YAL
 
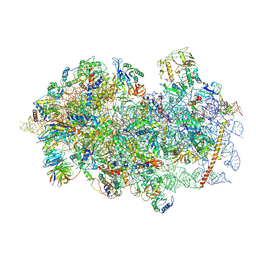 | | Mammalian 48S late-stage initiation complex with beta-globin mRNA | | 分子名称: | 18S ribosomal RNA, 40S Ribosomal protein uS3, 40S Ribosomal protein uS7, ... | | 著者 | Bochler, A, Simonetti, A, Guca, E, Hashem, Y. | | 登録日 | 2020-03-12 | | 公開日 | 2020-04-08 | | 最終更新日 | 2020-04-22 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural Insights into the Mammalian Late-Stage Initiation Complexes.
Cell Rep, 31, 2020
|
|
6XIJ
 
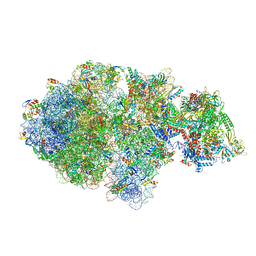 | | Escherichia coli transcription-translation complex A (TTC-A) containing an 24 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | 登録日 | 2020-06-20 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (8 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6XGF
 
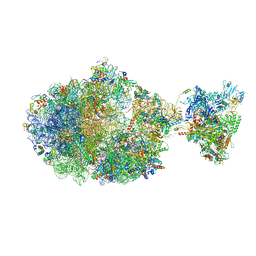 | | Escherichia coli transcription-translation complex B (TTC-B) containing an 30 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | 登録日 | 2020-06-17 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (5 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6XII
 
 | | Escherichia coli transcription-translation complex B (TTC-B) containing an 24 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | 登録日 | 2020-06-20 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (7 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
3JZU
 
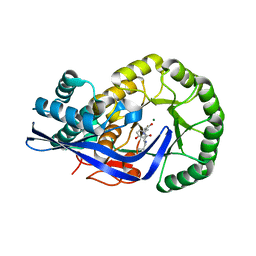 | | Crystal structure of Dipeptide Epimerase from Enterococcus faecalis V583 complexed with Mg and dipeptide L-Leu-L-Tyr | | 分子名称: | Dipeptide Epimerase, LEUCINE, MAGNESIUM ION, ... | | 著者 | Fedorov, A.A, Fedorov, E.V, Imker, H.J, Sakai, A, Gerlt, J.A, Almo, S.C. | | 登録日 | 2009-09-24 | | 公開日 | 2010-08-18 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7T82
 
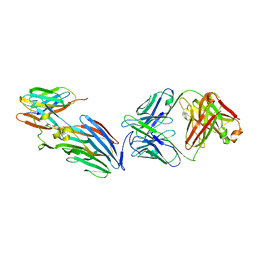 | | Crystal Structure of LEUKOCIDIN E/CENTYRIN S26/FAB B438 | | 分子名称: | Antibody Fab Heavy Chain, Antibody Fab Light Chain, Centyrin S26, ... | | 著者 | Luo, J, Malia, T.J, Buckley, P.T. | | 登録日 | 2021-12-15 | | 公開日 | 2023-01-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Multivalent human antibody-centyrin fusion protein to prevent and treat Staphylococcus aureus infections.
Cell Host Microbe, 31, 2023
|
|
7UVX
 
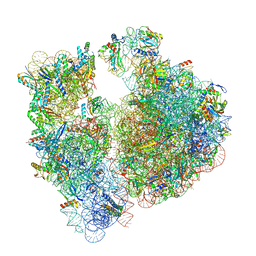 | | A. baumannii 70S ribosome-Streptothricin-F complex | | 分子名称: | 16s Ribosomal RNA, 23s ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Morgan, C.E, Yu, E.W. | | 登録日 | 2022-05-02 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.35 Å) | | 主引用文献 | Streptothricin F is a bactericidal antibiotic effective against highly drug-resistant gram-negative bacteria that interacts with the 30S subunit of the 70S ribosome.
Plos Biol., 21, 2023
|
|
7UVY
 
 | |
7UVV
 
 | |
2HG0
 
 | | Structure of the West Nile Virus envelope glycoprotein | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein | | 著者 | Nybakken, G.E, Nelson, C.A, Chen, B.R, Diamond, M.S, Fremont, D.H. | | 登録日 | 2006-06-26 | | 公開日 | 2006-11-07 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structure of the West Nile virus envelope glycoprotein.
J.Virol., 80, 2006
|
|
6X7F
 
 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B2 (TTC-B2) containing an mRNA with a 24 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | 登録日 | 2020-05-29 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6XDQ
 
 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B3 (TTC-B3) containing an mRNA with a 30 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | 登録日 | 2020-06-11 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
4R5P
 
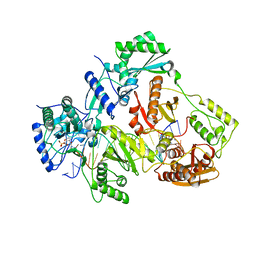 | | Crystal structure of HIV-1 reverse transcriptase (RT) with DNA and a nucleoside triphosphate mimic alpha-carboxy nucleoside phosphonate inhibitor | | 分子名称: | 5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*G)-3', 5'-D(*TP*GP*GP*AP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3', HIV-1 reverse transcriptase, ... | | 著者 | Das, K, Martinez, S.E, Arnold, E. | | 登録日 | 2014-08-21 | | 公開日 | 2015-03-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.894 Å) | | 主引用文献 | Alpha-carboxy nucleoside phosphonates as universal nucleoside triphosphate mimics.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6G2T
 
 | | human cardiac myosin binding protein C C1 Ig-domain bound to native cardiac thin filament | | 分子名称: | Actin, cytoplasmic 2, Myosin-binding protein C, ... | | 著者 | Risi, C, Belknap, B, Forgacs, E, Harris, S.P, Schroder, G.F, White, H.D, Galkin, V.E. | | 登録日 | 2018-03-23 | | 公開日 | 2018-10-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (9 Å) | | 主引用文献 | N-Terminal Domains of Cardiac Myosin Binding Protein C Cooperatively Activate the Thin Filament.
Structure, 26, 2018
|
|
7MM6
 
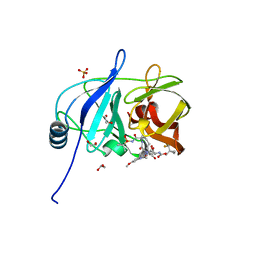 | |
7MM8
 
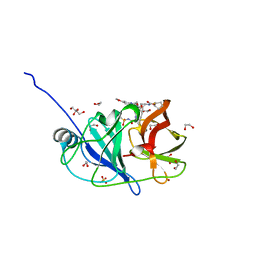 | | Crystal structure of HCV NS3/4A protease in complex with NR02-08 | | 分子名称: | (1R,2R)-2-fluorocyclopentyl {(2R,4S,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, 1,2-ETHANEDIOL, NS3/4a protease, ... | | 著者 | Zephyr, J, Schiffer, C.A. | | 登録日 | 2021-04-29 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MM2
 
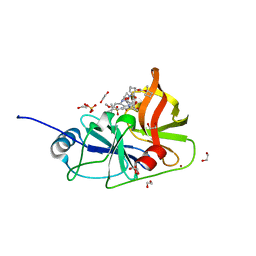 | | Crystal structure of HCV NS3/4A protease in complex with NR02-61 | | 分子名称: | 1,2-ETHANEDIOL, 1-methylcyclobutyl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, NS3/4a protease, ... | | 著者 | Zephyr, J, Schiffer, C.A. | | 登録日 | 2021-04-29 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.891 Å) | | 主引用文献 | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MMJ
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR02-08 | | 分子名称: | (1R,2R)-2-fluorocyclopentyl {(2R,4S,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, 1,2-ETHANEDIOL, NS3 protease, ... | | 著者 | Zephyr, J, Schiffer, C.A. | | 登録日 | 2021-04-29 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.992 Å) | | 主引用文献 | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MMF
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR02-60 | | 分子名称: | (1R)-1-cyclopentyl-2,2,2-trifluoroethyl {(2R,4R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, 1,2-ETHANEDIOL, NS3/4A protease, ... | | 著者 | Zephyr, J, Schiffer, C.A. | | 登録日 | 2021-04-29 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.891 Å) | | 主引用文献 | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
8APP
 
 | | AbLys1 endolysin from Acinetobacter baumannii phage AbTZA1 | | 分子名称: | Endolysin, GLYCEROL, PHOSPHATE ION | | 著者 | Premetis, G.E, Stathi, A, Papageorgiou, A.C, Labrou, N.E. | | 登録日 | 2022-08-10 | | 公開日 | 2022-12-07 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Characterization of a glycoside hydrolase endolysin from Acinetobacter baumannii phage AbTZA1 with high antibacterial potency and novel structural features.
Febs J., 290, 2023
|
|
7MM7
 
 | | Crystal structure of HCV NS3/4A protease in complex with NR02-23 | | 分子名称: | (2S)-1,1,1-trifluoropropan-2-yl {(2R,4S,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, 1,2-ETHANEDIOL, NS3/4a protease, ... | | 著者 | Zephyr, J, Schiffer, C.A. | | 登録日 | 2021-04-29 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.862 Å) | | 主引用文献 | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MM5
 
 | | Crystal structure of HCV NS3/4A protease in complex with NR02-60 | | 分子名称: | (1R)-1-cyclopentyl-2,2,2-trifluoroethyl {(2R,4R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, 1,2-ETHANEDIOL, NS3/4a protease, ... | | 著者 | Zephyr, J, Schiffer, C.A. | | 登録日 | 2021-04-29 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MMG
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR02-58 | | 分子名称: | 1-(trifluoromethyl)cyclobutyl {(2R,4S,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, ARGININE, NS3/4A protease, ... | | 著者 | Zephyr, J, Schiffer, C.A. | | 登録日 | 2021-04-29 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
