5Q1L
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A in complex with FMOPL000073a | | 分子名称: | (6~{R})-6-(4-methoxyphenyl)-2-oxidanylidene-5,6-dihydro-1~{H}-pyrimidine-4-carboxylic acid, DNA cross-link repair 1A protein, MALONATE ION, ... | | 著者 | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | 登録日 | 2017-05-15 | | 公開日 | 2018-08-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | PanDDA analysis group deposition
To Be Published
|
|
5PNZ
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 in complex with N10162a | | 分子名称: | 1,2-ETHANEDIOL, 1-[(4-methoxyphenyl)methyl]-1H-tetrazole, Bromodomain-containing protein 1, ... | | 著者 | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | 登録日 | 2017-02-07 | | 公開日 | 2017-03-15 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.557 Å) | | 主引用文献 | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5OLC
 
 | |
5O3K
 
 | |
5OW9
 
 | | Vitamin D receptor complex | | 分子名称: | (1~{S},3~{Z})-3-[(2~{E})-2-[(1~{S},3~{a}~{S},7~{a}~{S})-7~{a}-methyl-1-[(2~{S})-6-methyl-2-oxidanyl-heptan-2-yl]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexan-1-ol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | 著者 | Rochel, N, Li, W. | | 登録日 | 2017-08-31 | | 公開日 | 2018-02-07 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.403 Å) | | 主引用文献 | Investigation of 20S-hydroxyvitamin D3 analogs and their 1 alpha-OH derivatives as potent vitamin D receptor agonists with anti-inflammatory activities.
Sci Rep, 8, 2018
|
|
5NUR
 
 | | Structural basis for maintenance of bacterial outer membrane lipid asymmetry | | 分子名称: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)-2-amino-2-deoxy-4-O-phosphono-beta-D-glucopyranose-(1-6)-2-amino-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(3-4)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)-2-amino-2-deoxy-4-O-phosphono-beta-D-glucopyranose-(1-6)-2-amino-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, ... | | 著者 | Abellon-Ruiz, J, Kaptan, S.S, Basle, A, Claudi, B, Bumann, D, Kleinekathofer, U, van den Berg, B. | | 登録日 | 2017-05-01 | | 公開日 | 2017-10-25 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.29 Å) | | 主引用文献 | Structural basis for maintenance of bacterial outer membrane lipid asymmetry.
Nat Microbiol, 2, 2017
|
|
5NYV
 
 | | Crystal structure determination from picosecond infrared laser ablated protein crystals by serial synchrotron crystallography | | 分子名称: | Fluoroacetate dehalogenase | | 著者 | Schulz, E.C, Kaub, J, Busse, F, Mehrabi, P, Mueller-Werkmeiser, H, Pai, E.F, Robertson, W.D, Miller, R.J.D. | | 登録日 | 2017-05-11 | | 公開日 | 2018-03-21 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Protein crystals IR laser ablated from aqueous solution at high speed retain their diffractive properties: applications in high-speed serial crystallography.
J.Appl.Crystallogr., 50, 2017
|
|
5O04
 
 | |
5OCV
 
 | | A Rare Lysozyme Crystal Form Solved Using High-Redundancy 3D Electron Diffraction Data from Micron-Sized Needle Shaped Crystals | | 分子名称: | Lysozyme C, SODIUM ION | | 著者 | Xu, H, Lebrette, H, Yang, T, Srinivas, V, Hovmoller, S, Hogbom, M, Zou, X. | | 登録日 | 2017-07-03 | | 公開日 | 2018-03-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (2.2 Å) | | 主引用文献 | A Rare Lysozyme Crystal Form Solved Using Highly Redundant Multiple Electron Diffraction Datasets from Micron-Sized Crystals.
Structure, 26, 2018
|
|
5O02
 
 | |
5NY5
 
 | | The apo structure of 3,4-dihydroxybenzoic acid decarboxylases from Enterobacter cloacae | | 分子名称: | 3,4-dihydroxybenzoate decarboxylase, GLYCEROL | | 著者 | Dordic, A, Gruber, K, Payer, S, Glueck, S, Pavkov-Keller, T, Marshall, S, Leys, D. | | 登録日 | 2017-05-11 | | 公開日 | 2017-09-13 | | 最終更新日 | 2020-11-18 | | 実験手法 | X-RAY DIFFRACTION (2.501 Å) | | 主引用文献 | Regioselective para-Carboxylation of Catechols with a Prenylated Flavin Dependent Decarboxylase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5NZU
 
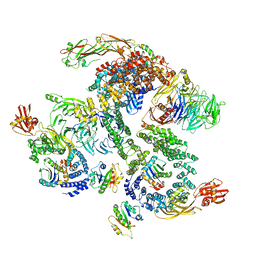 | | The structure of the COPI coat linkage II | | 分子名称: | ADP-ribosylation factor 1, Coatomer subunit alpha, Coatomer subunit beta, ... | | 著者 | Dodonova, S.O, Aderhold, P, Kopp, J, Ganeva, I, Roehling, S, Hagen, W.J.H, Sinning, I, Wieland, F, Briggs, J.A.G. | | 登録日 | 2017-05-15 | | 公開日 | 2017-06-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (15 Å) | | 主引用文献 | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
5Q0N
 
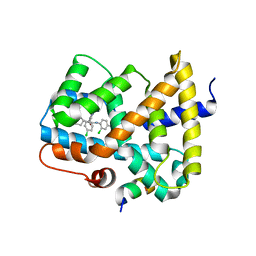 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | 3-chloro-4-({(2S)-2-[2-(4-chlorophenyl)-5,6-difluoro-1H-benzimidazol-1-yl]-2-cyclohexylacetyl}amino)benzoic acid, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2021-11-17 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q10
 
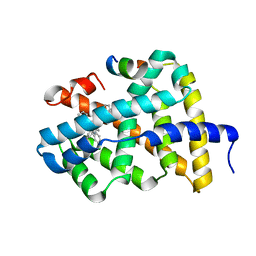 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, N-[3-(acetylamino)phenyl]-4-chloro-N-[(1S)-1-cyclohexyl-2-(cyclohexylamino)-2-oxoethyl]benzamide | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q1D
 
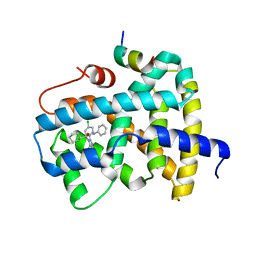 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | (2S)-2-[2-(4-chlorophenyl)-5,6-difluoro-1H-benzimidazol-1-yl]-2-cyclohexyl-N-phenylacetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0J
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | (2S)-N,2-dicyclohexyl-2-[2-(5-phenylthiophen-2-yl)-1H-benzimidazol-1-yl]acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2021-11-17 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0X
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | 6-(4-{[3-(3,5-dichloropyridin-4-yl)-5-(1-methylethyl)isoxazol-4-yl]methoxy}-2-methylphenyl)-1-methyl-1H-indole-3-carbox ylic acid, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q15
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | (2S)-N,2-dicyclohexyl-2-{5,6-difluoro-2-[(R)-methoxy(phenyl)methyl]-1H-benzimidazol-1-yl}acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0T
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | 2-phenyl-N-(propan-2-yl)-6-[(thiophen-2-yl)sulfonyl]-4,5,6,7-tetrahydro-1H-pyrrolo[2,3-c]pyridine-1-carboxamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q11
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, N,N-dicyclohexyl-3-(2,4-dichlorophenyl)-5-methyl-1,2-oxazole-4-carboxamide | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q18
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | (2S)-2-cyclohexyl-2-{5,6-difluoro-2-[(R)-methoxy(phenyl)methyl]-1H-benzimidazol-1-yl}-N-(trans-4-hydroxycyclohexyl)acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
7RZW
 
 | | CryoEM structure of Arabidopsis thaliana phytochrome B | | 分子名称: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome B | | 著者 | Li, H, Burgie, E.S, Vierstra, R.D, Li, H. | | 登録日 | 2021-08-27 | | 公開日 | 2022-04-13 | | 最終更新日 | 2022-04-20 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Plant phytochrome B is an asymmetric dimer with unique signalling potential.
Nature, 604, 2022
|
|
7SCG
 
 | | FH210 bound Mu Opioid Receptor-Gi Protein Complex | | 分子名称: | (2E)-N-[(2S)-2-(dimethylamino)-3-(4-hydroxyphenyl)propyl]-3-(naphthalen-1-yl)prop-2-enamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Wang, H, Kobilka, B. | | 登録日 | 2021-09-28 | | 公開日 | 2022-04-20 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structure-Based Evolution of G Protein-Biased mu-Opioid Receptor Agonists.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7S76
 
 | | HBV CAPSID Y132A WITH COMPOUND 10b AT 2.5A RESOLUTION | | 分子名称: | (1-methyl-1H-1,2,4-triazol-3-yl)methyl {(4S)-1-[(3-chloro-4-fluorophenyl)carbamoyl]-2-methyl-2,4,5,6-tetrahydrocyclopenta[c]pyrrol-4-yl}carbamate, Capsid protein | | 著者 | Olland, A.M, Suto, R.K. | | 登録日 | 2021-09-15 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The identification of highly efficacious functionalised tetrahydrocyclopenta[ c ]pyrroles as inhibitors of HBV viral replication through modulation of HBV capsid assembly.
Rsc Med Chem, 13, 2022
|
|
7SIT
 
 | | Crystal structure of Voltage gated potassium ion channel, Kv 1.2 chimera-3m | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OXYGEN ATOM, POTASSIUM ION, ... | | 著者 | Reddi, R, Matulef, K, Riederer, E.A, Whorton, M.R, Valiyaveetil, F.I. | | 登録日 | 2021-10-14 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.32 Å) | | 主引用文献 | Structural basis for C-type inactivation in a Shaker family voltage-gated K + channel.
Sci Adv, 8, 2022
|
|
